Figure 1. Secondary and tertiary structural analysis of the proteins. A: The backbone folding does not alter upon mutation. Estimation of the secondary structures of the proteins, using far-ultraviolet
circular dichroism (CD) spectra in the region 195–250 nm, using a JASCO CD spectrometer, at ambient temperature (27 °C), recorded
with 2 mm path length cells. B: Intrinsic fluorescence of the wild-type and D26G human gamma S-crystallin (HGSC) differ little: Curve A (Blue): γS wild-type; B (Red): D26G; with excitation wavelength 295 nm and emission wavelength recorded from 300 to 400 nm, at ambient temperature,
using a Hitachi spectrofluorimeter. C: The mutation has a slightly more set of surface-exposed residues: Extrinsic emission spectra of the surface probe Nile Red.
A (Blue): γS wild-type; B (Red): D26G; extrinsic fluorescence spectra were recorded between 570 and 700 nm with excitation at 540 nm; and slit size
10 nm for excitation and emission. In each set of spectra in A, B, and C, the protein concentrations used were 5 µM (0.1 mg/ml) in 50 mM Tris buffer (pH 7.3), cell path length 3 mm, and spectra
recorded at 5.0 nm excitation and emission slits. Spectra shown were the average of three runs.
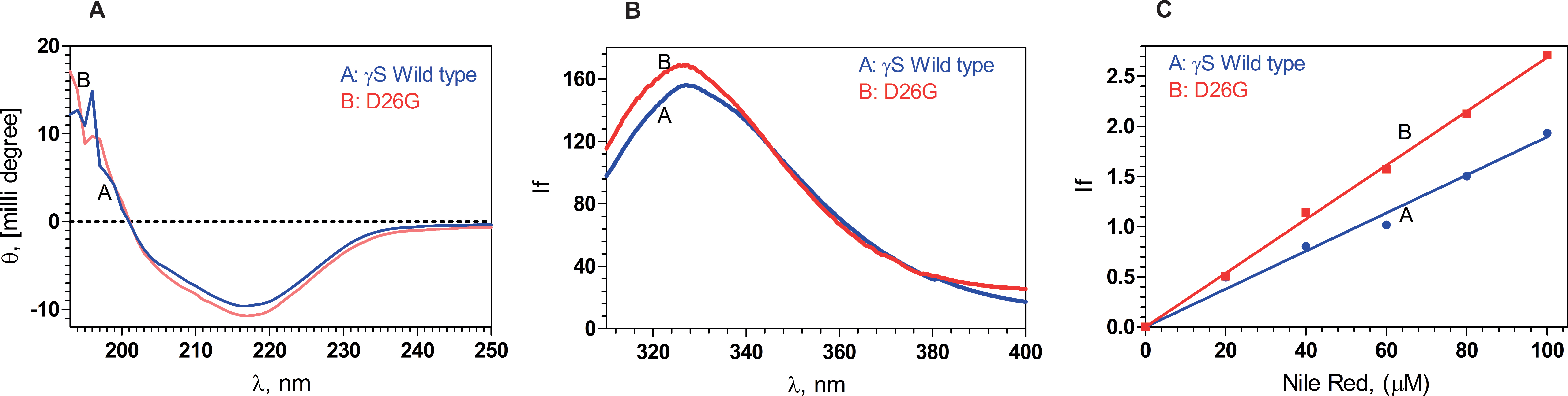
 Figure 1 of
Karri, Mol Vis 2013; 19:1231-1237.
Figure 1 of
Karri, Mol Vis 2013; 19:1231-1237.  Figure 1 of
Karri, Mol Vis 2013; 19:1231-1237.
Figure 1 of
Karri, Mol Vis 2013; 19:1231-1237. 