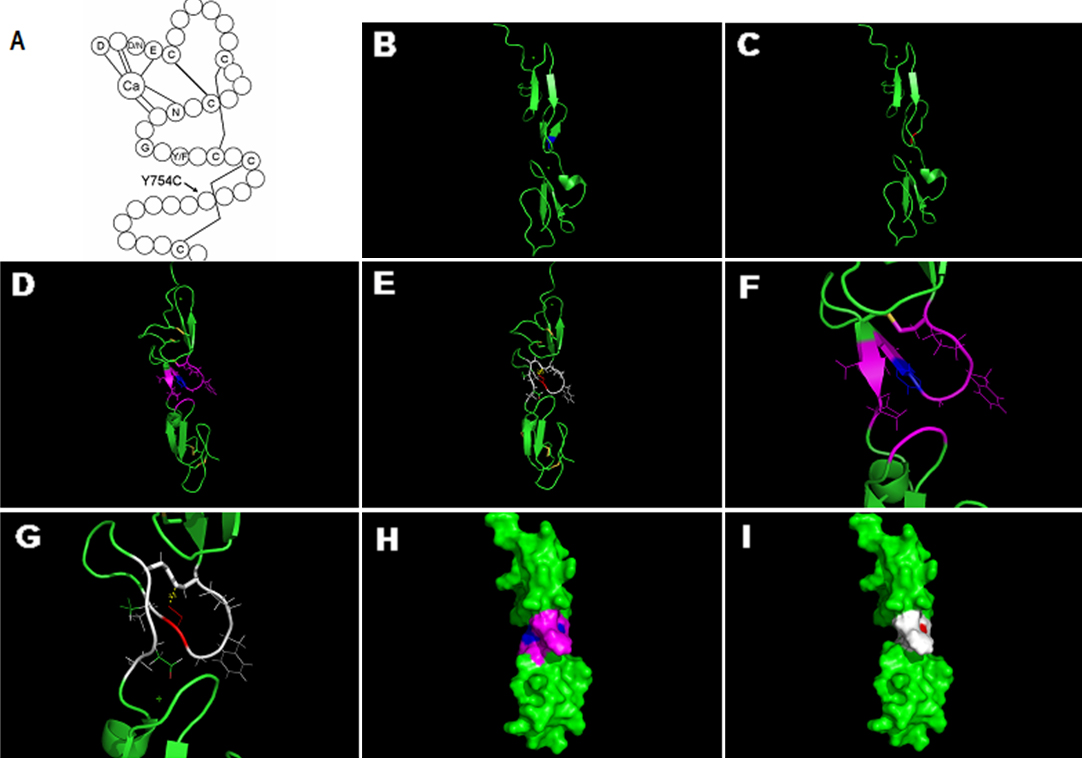
Figure 4. Structure analyses of the missense mutation in the calcium binding (cb) epidermal growth factor (EGF)-like7 domain.
A: The consensus secondary structure of a prototypical cbEGF-like domain. Calcium binding in the NH
2-terminal region of the wild-type domain is mediated by the consensus sequence (D/N) -X- (D/N) (E/Q) Xm (D/N) Xn (Y/F; m and
n are variables), and highly conserved amino acids are identified by their single-letter amino acid code. The letter C in
the schematic represents the highly conserved cysteine of cbEGF-like domain, and the lines between cysteine represent disulfide
bridges. The mutation p.Y754C located at the region between the last two cysteines of the domain, which could probably interfere
with the disulfide bond formation between the two cysteines.
B: The 3D structure of the wild cbEGF-like7–8 domains, which are created based on the Protein Data Bank (PDB) template 1EMN
(47% sequence identity) by
PyMOL 1.1r1. The blue represents the unaffected tyrosine.
C: The potential conformation change of the mutation. The red represents the substitute cysteine, where the double β-sheet
transformed to a loop-region.
D and
E: The 3D structure of domains in
B and
C, respectively. The yellow lines represent disulfide bonds, and the blue represents the unaffected tyrosine. The purple displays
the residues within the distance of 4Å with tyrosine
754, and double β-sheet between obligatory glycine
753 and aspartic
765. The red represents the substitute cysteine, which absents the benzene ring. The yellow dashed line represents the potential
disulfide bond formation between the introduced cysteine
754 and cysteine
750, which would probably disrupt the disulfide bond between cysteine
750 and cysteine
763. The white displays the residues within the distance of 4Å with cysteine
754; compared with the purple wild type, β-turn, which forms the calcium binding pocket of cbEGF-like8, is farther in distance
with cysteine
754.
F and
G: A zoom-in change of
D and
E, respectively.
H and
I: The surface of the wild and mutant cbEGF-like7–8 domains, respectively. The colors correspond to that of figure
D and
E. The surface area of the mutant region is smaller than that of the wild. In summary, the conformation of the mutant domain
is likely to be strongly altered, to include neighboring domain as well.
 Figure 4 of
Li, Mol Vis 2012; 18:504-511.
Figure 4 of
Li, Mol Vis 2012; 18:504-511.  Figure 4 of
Li, Mol Vis 2012; 18:504-511.
Figure 4 of
Li, Mol Vis 2012; 18:504-511.