Figure 2. Results of sequencing and polymerase chain reaction (PCR)-fragment length polymorphism (PCR-FLP) analyses for the patient
with FCD are demonstrated. A: Nucleotide and amino acid sequence of the wild (upper) and mutated (lower) PIKFYVE gene around the identified p.Glu1389AspfsX16 mutation are demonstrated. The deleted four bases of the c.4166_4169delAAGT
mutation are indicated in bold type in the wild-type sequence. The altered amino acid sequence downstream of the deleted four
bases is indicated in bold italics in the mutated sequence. Asterisk (*) means an ochre (TAA) stop codon. B: Results of sequencing analysis for exon 24 of the PIKFYVE gene in normal volunteer (upper) and the patient with FCD (lower) from forward (left) and reverse (right) directions are
demonstrated. Arrowheads indicate the breakpoint of the c.4166_4169delAAGT mutation. C: The mixed base sequence (upper) downstream of the breakpoints was subtracted (middle) from the reference sequence (black
type) to extract the mutated sequence (red type) in both directions (left: forward, right: reverse). Note that the mutated
sequence is fully matched to the reference sequence from four bases downstream of the breakpoints (lower), indicating that
the mutated sequence is deleted with four bases, AAGT sequence. D: Results of PCR-FLP analysis for exon 24 of the PIKFYVE gene in normal volunteers (lanes 1 and 2) and the patient with FCD (lane 3) and her sister (lane 4) are demonstrated. Lane
5 means negative control. Note that the shorter PCR band in the patient with FCD (lane 3) was amplified from the mutated allele
while the longer PCR band was from the wild-type allele.
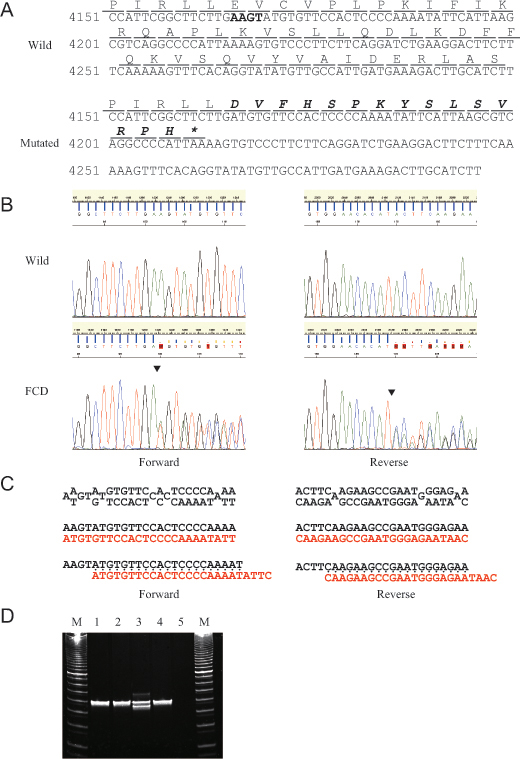
 Figure 2 of
Kawasaki, Mol Vis 2012; 18:2954-2960.
Figure 2 of
Kawasaki, Mol Vis 2012; 18:2954-2960.  Figure 2 of
Kawasaki, Mol Vis 2012; 18:2954-2960.
Figure 2 of
Kawasaki, Mol Vis 2012; 18:2954-2960. 