Figure 3. The analysis of potential microR-29b binding sites within the 3′-UTR of PKR associated protein X (RAX) mRNA. The bioinformatic algorithms TargetScan, miRanda,
and FindTar were used to predict miR-29 binding sites in the 3′-UTR of RAX mRNA. A: A map of the 3′-UTR of RAX mRNA was generated to indicate the putative binding sites, as predicted by the three bioinformatic algorithms. The seed region
of this common miRNA binding site is highly conserved among mammals. B: The diagram shows the target sites for the microRNA-29b (miR-29b) paralogs in the 3′- untranslated region (3′-UTR) of PKR associates protein X (RAX) mRNA, including the RNA hybrid-free energy
calculations and the theoretical miRNA-mRNA duplex pairing. The colon (:) represents the location of a G/U wobble base pairing.
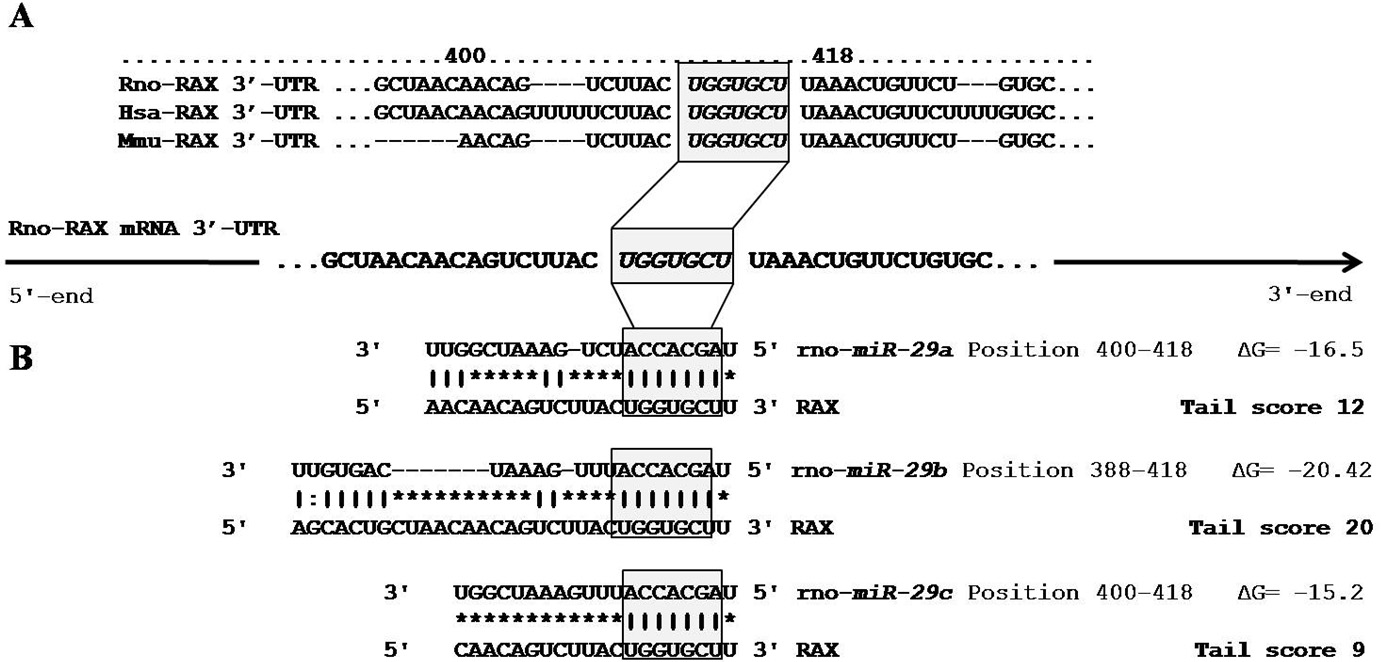
 Figure 3 of
Silva, Mol Vis 2011; 17:2228-2240.
Figure 3 of
Silva, Mol Vis 2011; 17:2228-2240.  Figure 3 of
Silva, Mol Vis 2011; 17:2228-2240.
Figure 3 of
Silva, Mol Vis 2011; 17:2228-2240. 