Figure 2. Homology modelling of K434R. A model of STI1's TPR2B domain (gray ribbon), based on homology to the TPR1 domain that was co-crystallized
with the COOH-terminal EEVD-containing peptide of HSP70 (PDB# 1elw_A), shows that residue 434 would not participate in binding
to a putative EEVD ligand (yellow ball-and-stick cluster). In silico mutagenesis of K434R predicts that there would also be
no increase of the global folding energy of the TPR2B domain. The seven alpha-helices of typical TPR domain structure are
labeled “A1”-”C.” The wild-type and K434R models were submitted to an Atomic Non Local Environment Assessment (ANOLEA) server
to compute folding energy; energy differences are in E/kT units, where E represents energy; k, the Boltzmann constant; and T, absolute temperature. Groups of residues around positions 420 and 434 are predicted to be
in a more energetically-favorable conformation in the K434R model.
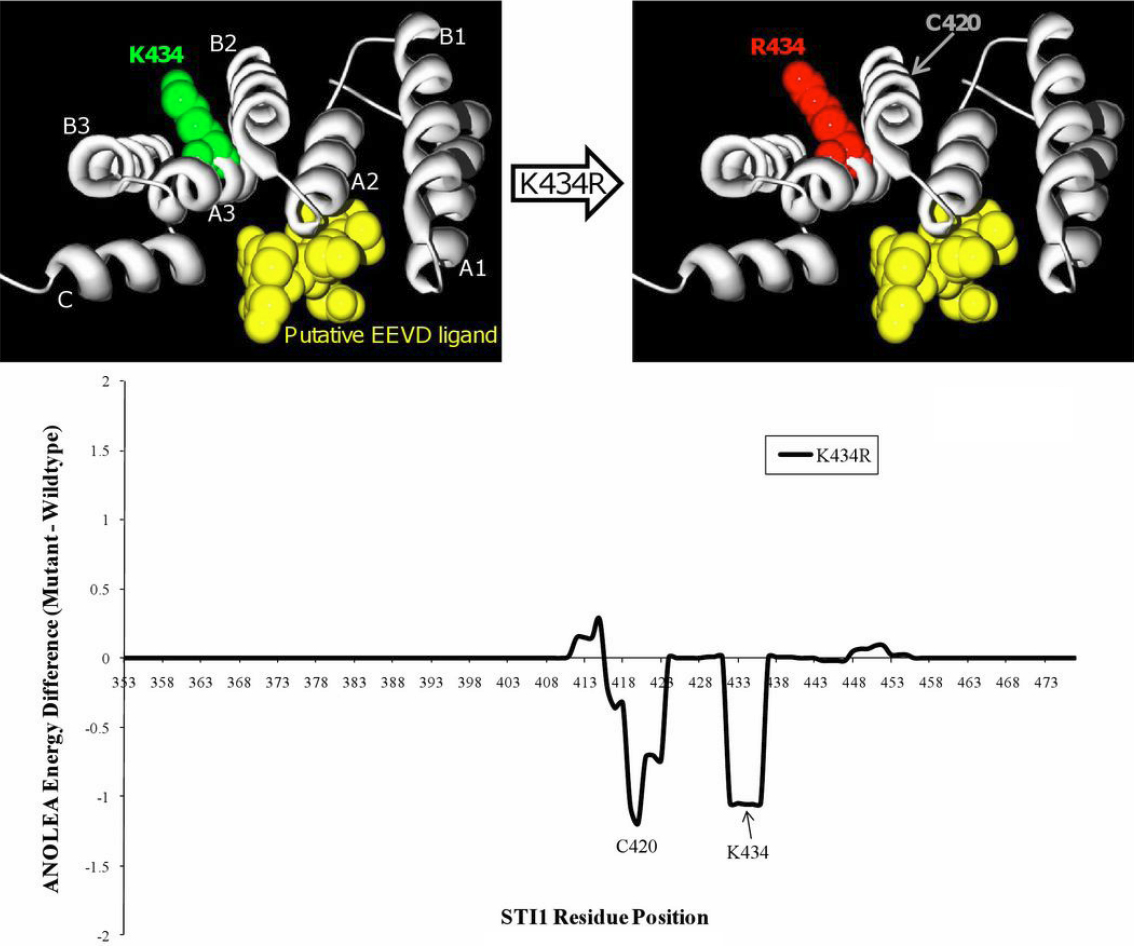
 Figure 2 of
Footz, Mol Vis 2011; 17:1957-1969.
Figure 2 of
Footz, Mol Vis 2011; 17:1957-1969.  Figure 2 of
Footz, Mol Vis 2011; 17:1957-1969.
Figure 2 of
Footz, Mol Vis 2011; 17:1957-1969. 