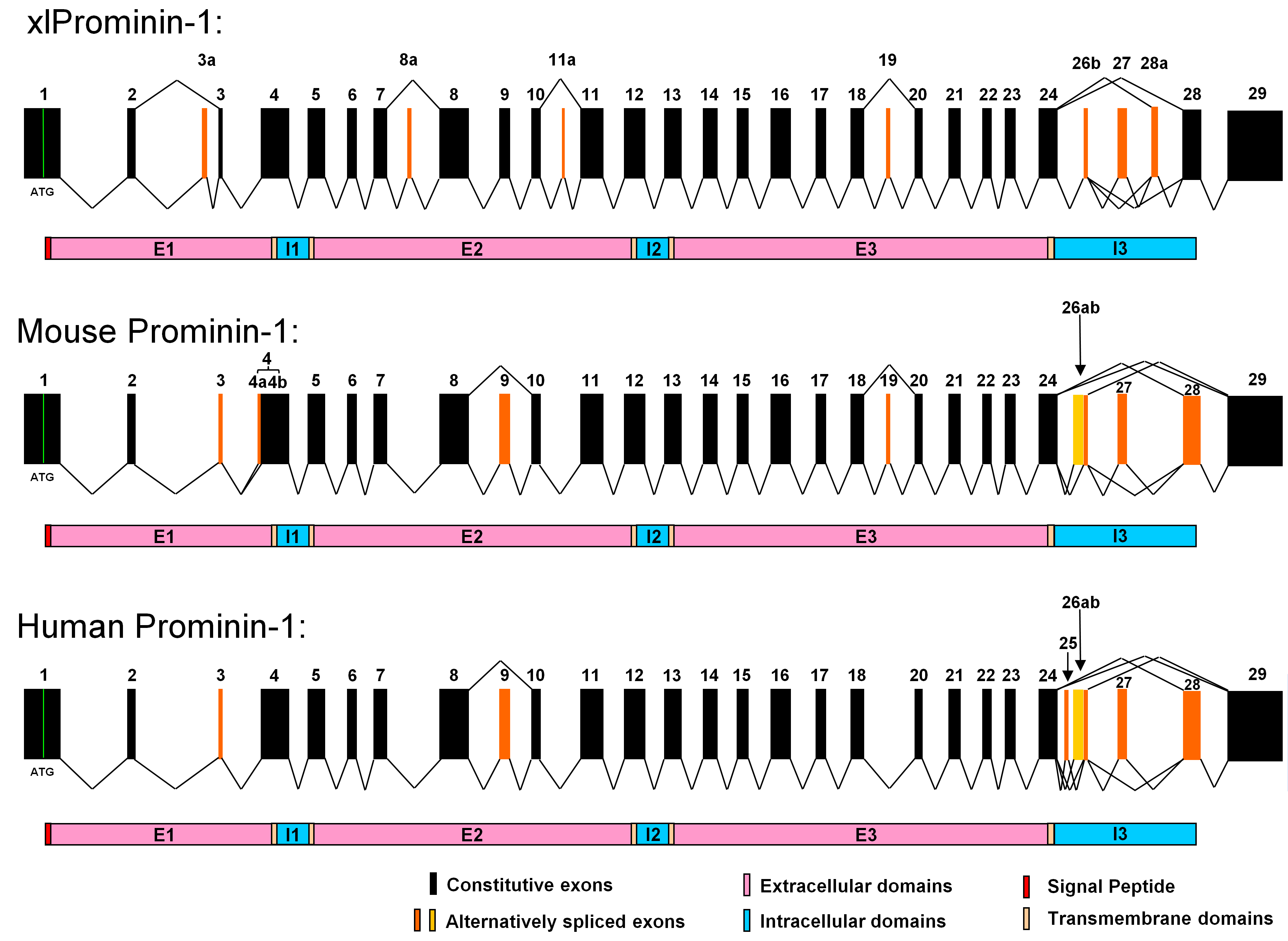
Figure 6. Comparison of exon organization
of prominin-1 gene from
X. laevis, mouse, and human. It appears
that the gene structure of prominin-1 is evolutionarily conserved in
these animals. Alternative splicing of the prominin-1 gene is seen in
all three animals, however, with considerable differences in the
choices of alternative exons and the splicing patterns. Homologous
exons of xlProminin-1 and mouse and human prominin-1 are aligned for
comparison of their exon organization. Constitutive exons are marked in
black. Alternatively spliced exons are marked in orange. Spliced forms
identified in cDNA clones are indicated by jointed lines. Note that
homologous exons are assigned with the same number to maintain
consistency with preexisting nomenclature [
2,
31].
Exons
26b and 27 are conserved and alternatively spliced in all three
species. Alternative exons 3a, 8a, 11a, and 28a of xlProminin-1 are not
found in mouse or human prominin-1. Splicing of alternative exons 4a
results from using an alternative 3′ splice site in exon 4, and is only
observed in mouse prominin-1 [
31].
Alternative
exon 19 of xlProminin-1 is alternatively spliced in mouse
prominin-1, but no evidence has been found that this exon is
alternatively spliced in human prominin-1. Alternative exon 25 of human
prominin-1 is not found in prominin-1 from the mouse or
X. laevis.
The
alternative exon 26a of mouse prominin-1 and the alternative exons
25 and 26a of human prominin-1 are not found in xlProminin-1. Exons 3,
9, and 28 of mouse prominin-1 are alternative, but appear to be
constitutive in
X. laevis. The region encompassing exons 24 to
28 of xlProminin-1 and homologous sequences of the prominin-1 gene from
the mouse and human are regions of extensive alternative splicing.
Diagrams of translated proteins are aligned with their coding
sequences. Divisions of proteins by predicted transmembrane domains are
marked.
 Figure 6 of Han, Mol Vis 2011; 17:1381-1396.
Figure 6 of Han, Mol Vis 2011; 17:1381-1396.  Figure 6 of Han, Mol Vis 2011; 17:1381-1396.
Figure 6 of Han, Mol Vis 2011; 17:1381-1396.