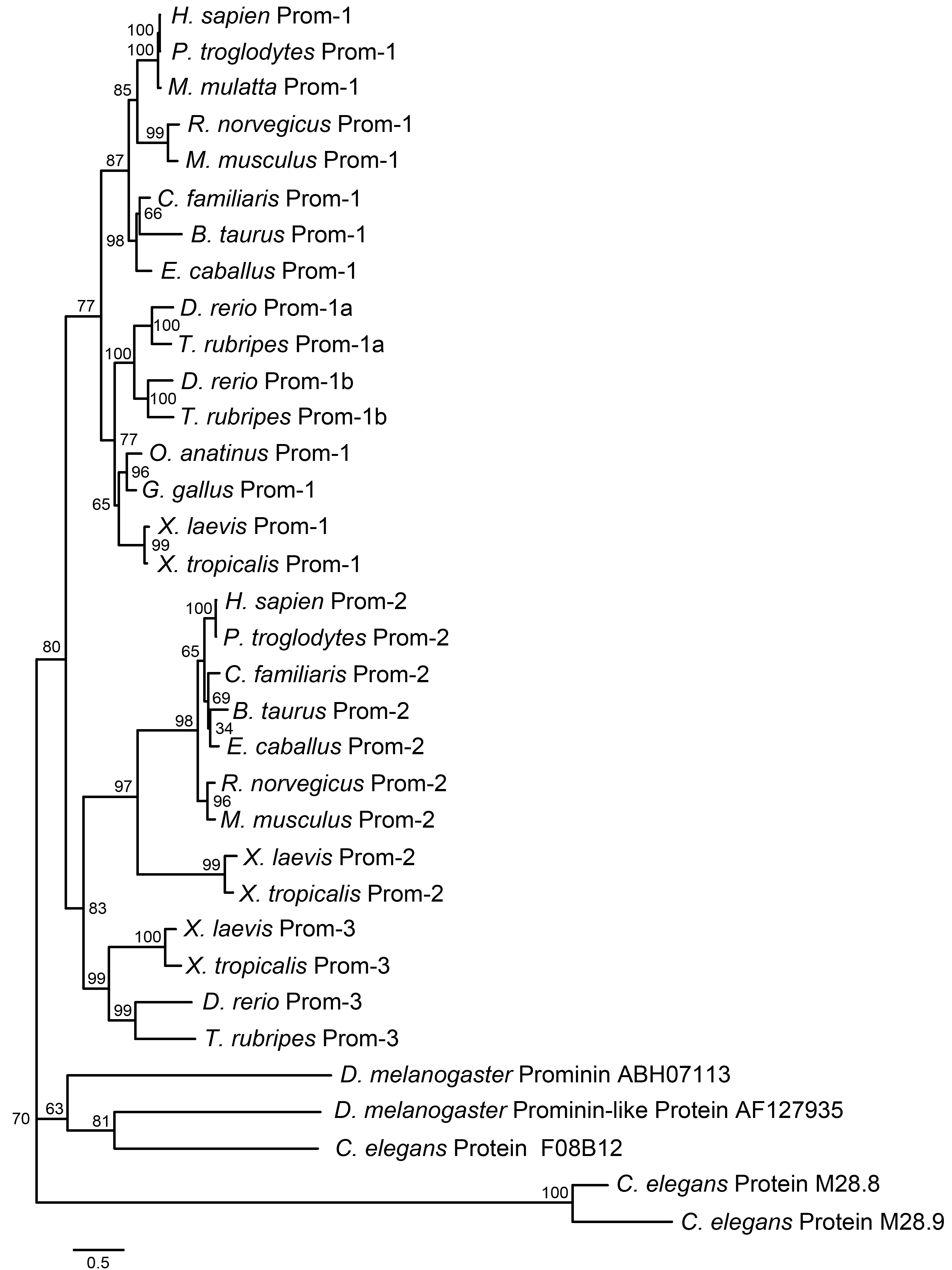
Figure 2. Phylogenic analysis of prominin
homologs from several species. This phylogenetic tree was constructed
to show the existence of multiple prominin homologs in metazoan animals
from fly to human and their evolutionary relationship. Three
X.
laevis prominin homologs, designated as xlProminin-1, 2, and 3, are
placed in branches of the consensus phylogenetic tree with prominin-1,
2, and 3 from other species. The numbers at each node are the
percentage bootstrapping values of 100 replicates. The evolutionary
distance of any two sequences is represented by the length of the
branches. Sequences used in the phylogenetic analysis are excerpted
from:
H. sapien Prom-1 (
NP_006008),
P.
troglodytes Prom-1 (
XP_517115),
M.
mulatta Prom-1 (
NP_001070888),
R. norvegicus Prom-1 (
NP_068519),
M.
musculus Prom-1 (
NP_032961),
C.
familiaris Prom-1 (
XP_850831),
B.
taurus Prom-1 (
HQ159409),
E.
caballus Prom-1 (
XP_001498729),
D. rerio Prom-1a (
NP_001108615),
T. rubripes Prom-1a (
HQ159405),
D.
rerio Prom-1b (
NP_932337),
T.
rubripes Prom-1b (
HQ159406),
O.
anatinus Prom-1 (
HQ159408),
G.
gallus Prom-1 (
XP_001232165),
X. laevis Prom-1 (
NP_001163920),
X. tropicalis Prom-1 (
HQ159400),
H.
sapien Prom-2 (
NP_653308),
P.
troglodytes Prom-2 (
XP_001143498),
C. familiaris Prom-2 (
XP_854455),
B.
taurus Prom-2 (
XP_599188),
E.
caballus Prom-2 (
XP_001494215),
R. norvegicus Prom-2 (
AAN63818),
M.
musculus Prom-2 (
NP_620089),
X.
laevis Prom-2 (
NP_001163922),
X. tropicalis Prom-2 (
HQ159402),
X.
laevis Prom-3 (
NP_001163921),
X. tropicalis Prom-3 (
HQ159403),
D.
rerio Prom-3 (
XP_684527),
T.
rubripes Prom-3 (
HQ159407),
D.
melanogaster Prominin (
ABH07113),
D.
melanogaster Prominin-like protein AF127935 (
NP_647770),
C.
elegans protein F08B12 (
NP_509907),
C.
elegans protein M28.8 (
NP_496294),
C.
elegans protein M28.9 (
NP_496296).
 Figure 2 of Han, Mol Vis 2011; 17:1381-1396.
Figure 2 of Han, Mol Vis 2011; 17:1381-1396.  Figure 2 of Han, Mol Vis 2011; 17:1381-1396.
Figure 2 of Han, Mol Vis 2011; 17:1381-1396.