Figure 1. Predicted secondary structures
of the wild type and the mutant βA3-crystallin. The following sequences
were used for secondary structures prediction: wild type (Wild_type);
glutamine that residues at position 85 is replaced by a glutamic acid
residue (Q85E); glutamine that residues at position 180 is replaced by
a glutamic acid residue (Q180E); βA3-crystallin double deamidated
mutant (Q85EQ180E); or glycine residue at position 91 is removed
(G91Del). The secondary structure types are designated as follows
(according to DSSP program): H=alpha helix; B=residue in isolated
beta-bridge; E=extended strand, participates in beta ladder; G=3-helix
(3/10 helix); I=5 helix (pi helix); T=hydrogen bonded turn; S=bend;
C=the rest.
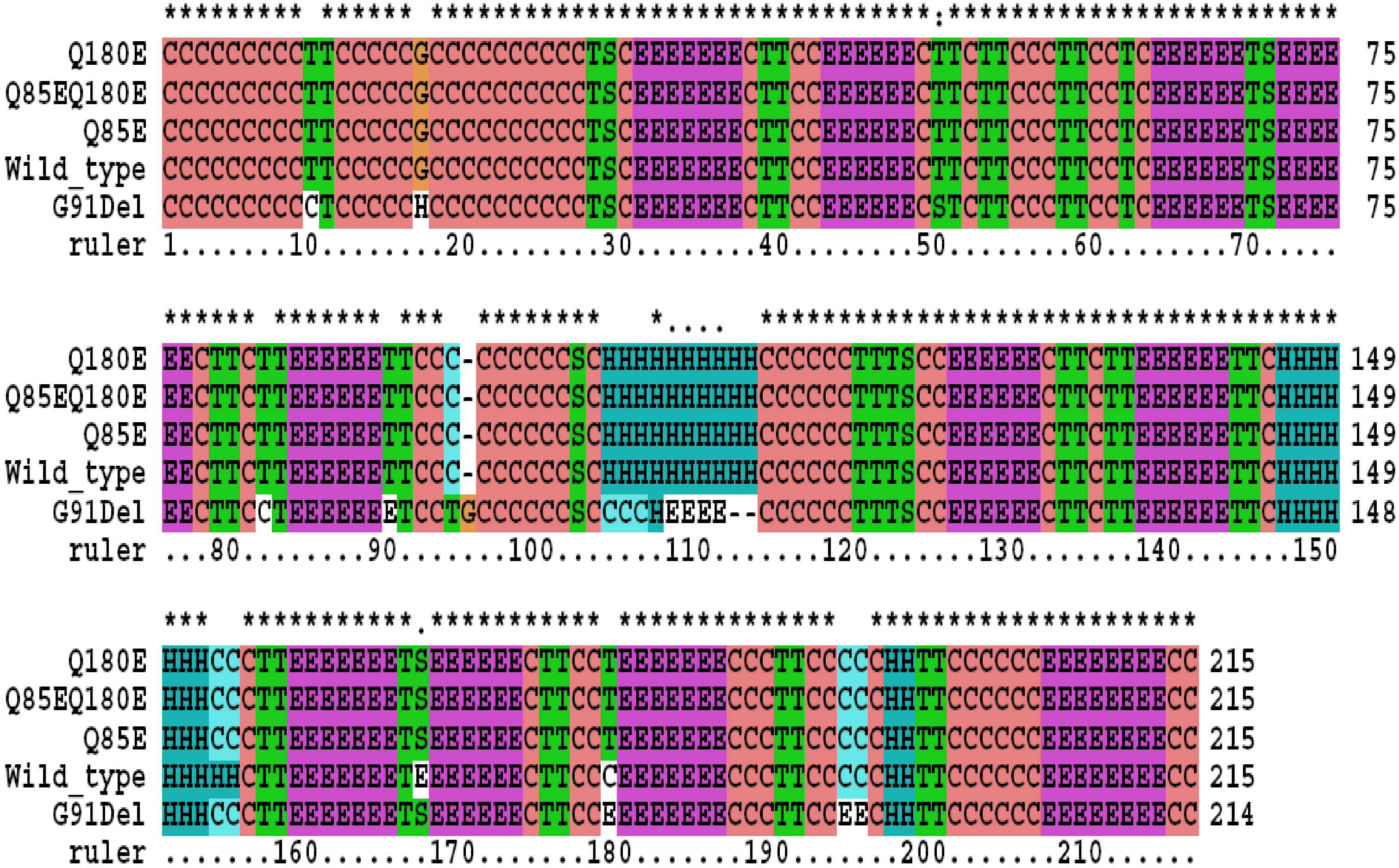
 Figure 1 of Xu, Mol Vis 2010; 16:438-444.
Figure 1 of Xu, Mol Vis 2010; 16:438-444.  Figure 1 of Xu, Mol Vis 2010; 16:438-444.
Figure 1 of Xu, Mol Vis 2010; 16:438-444. 