Figure 2. Result validation. The G32T
mutation was detected by direct sequencing in three Israeli family
members with HHCS. DNA samples from all three were also analyzed using
the MALDI-TOF technique and were used as positive controls for the G32T
mutation. The arrowhead indicates the unextended primer. The bold arrow
indicates the wild type allele. The thin arrow indicates the mutated
allele. Top panel: A spectrum from a template-free sample (negative
control). Middle panel: A spectrum from a patient negative for the
mutation. Bottom panel: A spectrum from a subject with HHCS (positive
control).
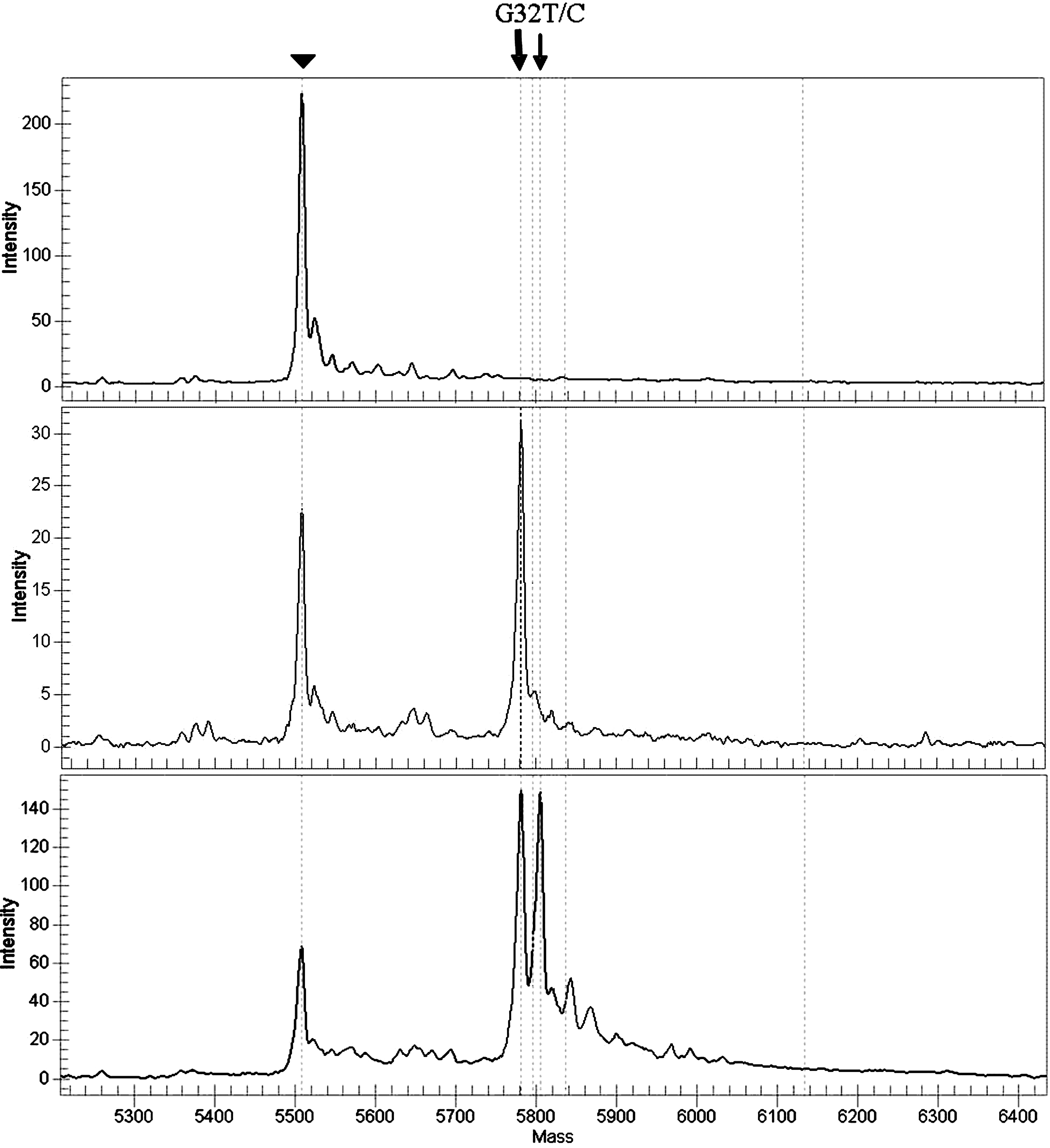
 Figure 2 of Assia, Mol Vis 2010; 16:2487-2493.
Figure 2 of Assia, Mol Vis 2010; 16:2487-2493.  Figure 2 of Assia, Mol Vis 2010; 16:2487-2493.
Figure 2 of Assia, Mol Vis 2010; 16:2487-2493. 