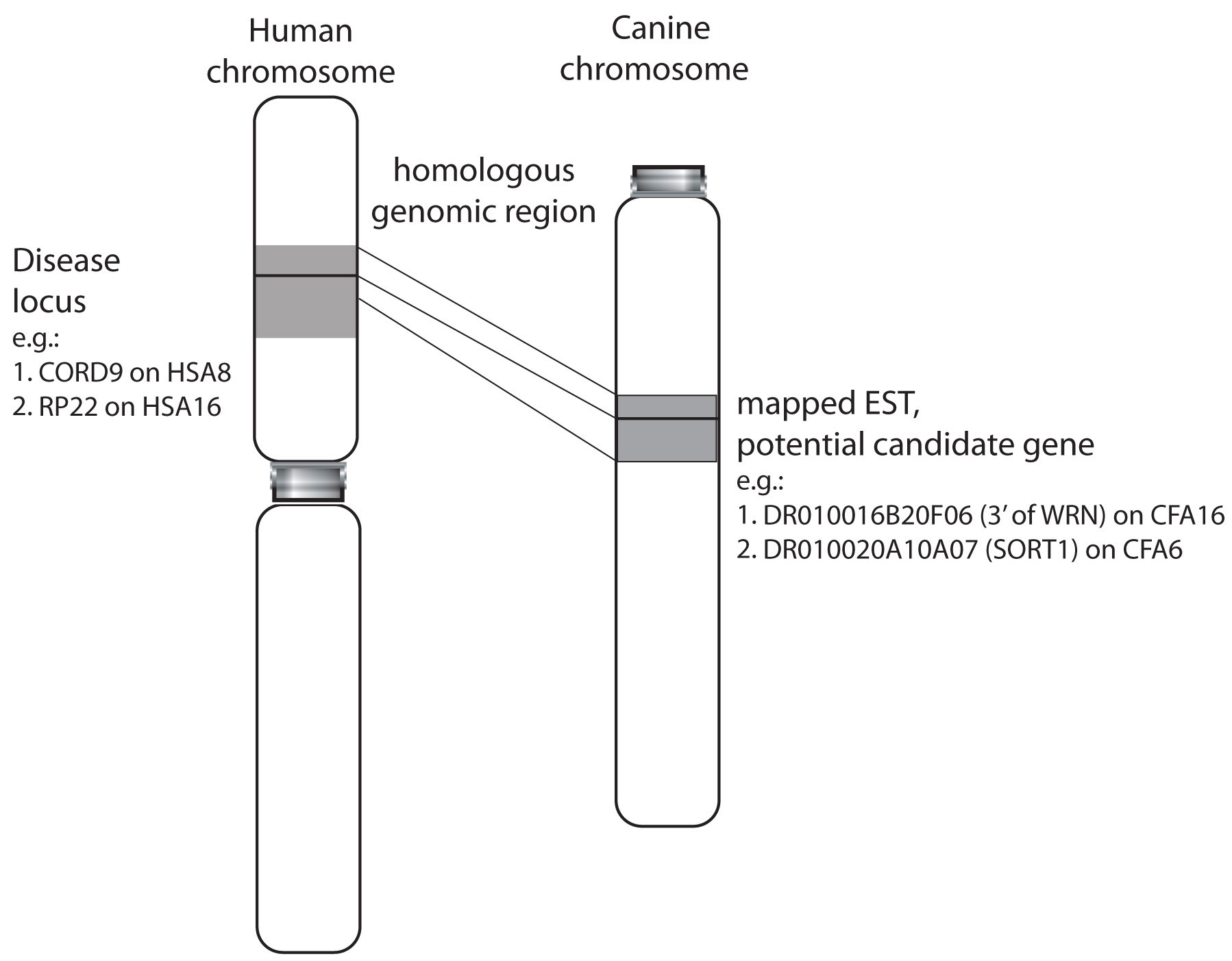
Figure 2. Identification of potential
candidate genes for human retinal disease Human genomic intervals for
known diseases (e.g., CORD9 on HSA8, RP22 on HSA16) were mapped against
the canine genome to identify homologous regions, and EST within these
regions of interest (e.g., DR010016B20F06 on CFA16, DR010020A10A07 on
CFA6). A comprehensive list of these disorders and the number of
corresponding ESTs contained within our library is given in
Appendix 2.
ESTs mapped in the presented research are also illustrated on the
respective chromosomes in
Appendix 5.
Details on all clones can be obtained through a web database (
DOG EST
or
DOG
EST Project) to obtain insights into corresponding transcripts
(e.g.: 1. WRN, 2. SORT1). We suggest that this tool provides new
positional candidate genes for mapped human retinal disorders. This
would allow for the identification of mutations in genes that are thus
far unknown or have not yet been linked to retinal disorders, after the
exclusion of conventional candidate genes.
 Figure 2 of Zangerl, Mol Vis 2009; 15:927-936.
Figure 2 of Zangerl, Mol Vis 2009; 15:927-936.  Figure 2 of Zangerl, Mol Vis 2009; 15:927-936.
Figure 2 of Zangerl, Mol Vis 2009; 15:927-936.