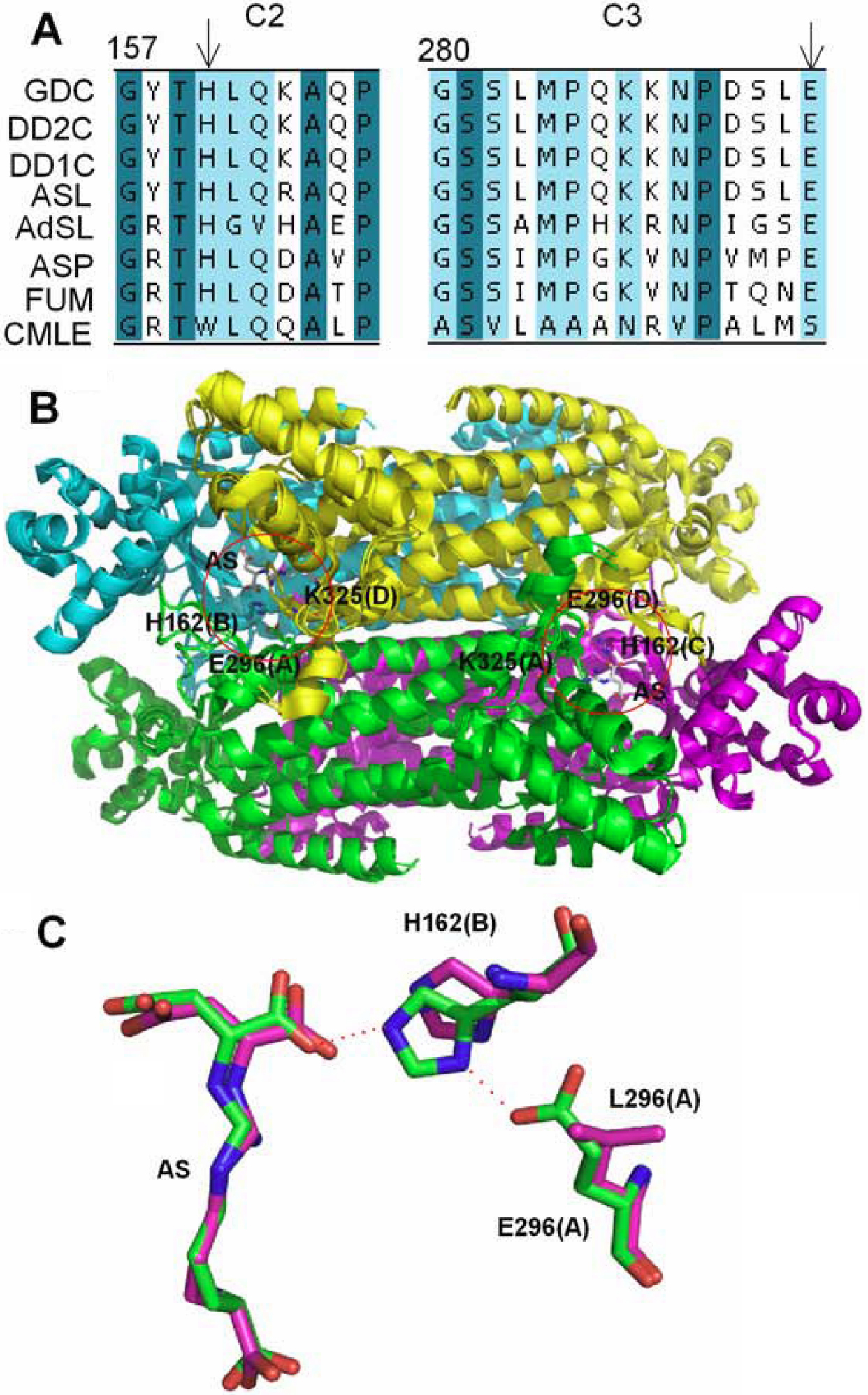
Figure 1. Comparison of sequence and
structure of δ–crystallin and the ASL super-family.
A:
Conserved regions of c2 (157–166) and c3 (280–294) are derived from
multiple-sequence alignment using the program ClustalW [
30]. Amino acid
sequences that are identical or highly conservative are shaded in dark
green and cyan, respectively. Abbreviations used: GDC, goose
δ–crystallin; DD2C, duck δ2–crystallin; DD1C, duck δ1–crystallin; ASL,
argininosuccinate lyase; FUM, fumarase; ASP, aspartase; CMLC, 3-carboxy-
cis-
cis-muconate
lactonizing enzyme. H160 and E294 are indicated by arrows.
B:
Superimposition of the quaternary structure of goose δ–crystallin and
duck T161D δ2–crystallin (PDB accession no: 1XWO and 1TJW). Monomer A,
B, C and D are shown as a cartoon and colored green, cyan, magenta and
yellow, respectively. The red circle depicts the active site. Related
residues are highlighted as sticks. Argininosuccinate (AS) is shown as
stick and carbon atoms are colored grey.
C: Superimposition of
residues in substrate binding site of duck T161D δ2–crystallin
structure and T161D/E296L δ2–crystallin model. E296, L296, H162 and AS
are displayed as stick models and shown in green and magenta for T161D
and T161D/E296L proteins, respectively. Interactions between E296 and
H162, and H162 and AS are highlighted by dashed orange lines. The
parentheses after each residue represent the defined subunit.
 Figure 1 of Huang, Mol Vis 2009; 15:2358-2363.
Figure 1 of Huang, Mol Vis 2009; 15:2358-2363.  Figure 1 of Huang, Mol Vis 2009; 15:2358-2363.
Figure 1 of Huang, Mol Vis 2009; 15:2358-2363.