Figure 3. Effect of NR2E3
mutations on DNA binding and transactivation. A: EMSAs were
performed using the [32P]-labeled Kni x2 probe with
untransfected (UnTR) COS-1 cells or WT NR2E3 expressing cell
extracts. Specificity of DNA binding is demonstrated by competition
with unlabeled Kni x2 oligonucleotide (50×) and nonspecific (NS)
oligonucleotide (50×). The arrow indicates the position of a specific
DNA–protein complex between NR2E3 and Kni x2 oligonucleotide. B:
Binding of mutant NR2E3 proteins to the labeled Kni x2 oligonucleotide
was examined by EMSA. Mutant NR2E3 protein amount in cell extracts was
normalized to WT-NR2E3 by immunoblot analysis. C: NR2E3
expression constructs were cotransfected into HEK293 cells with bovine
rhodopsin-130 luciferase reporter plasmid and with NRL and CRX
expression constructs. Fold change is relative to the empty expression
vector control. Error bars indicate standard error of mean (SE). D:
Luciferase assays were performed after co-transfection of mutant NR2E3
construct with the NRL and/or CRX expression constructs. ANOVA with a
post hoc test were performed on each sample compared to WT NR2E3.
Significant differences of p<0.05, and p<0.01 are shown as * and
**, respectively. Error bars correspond to SEM.
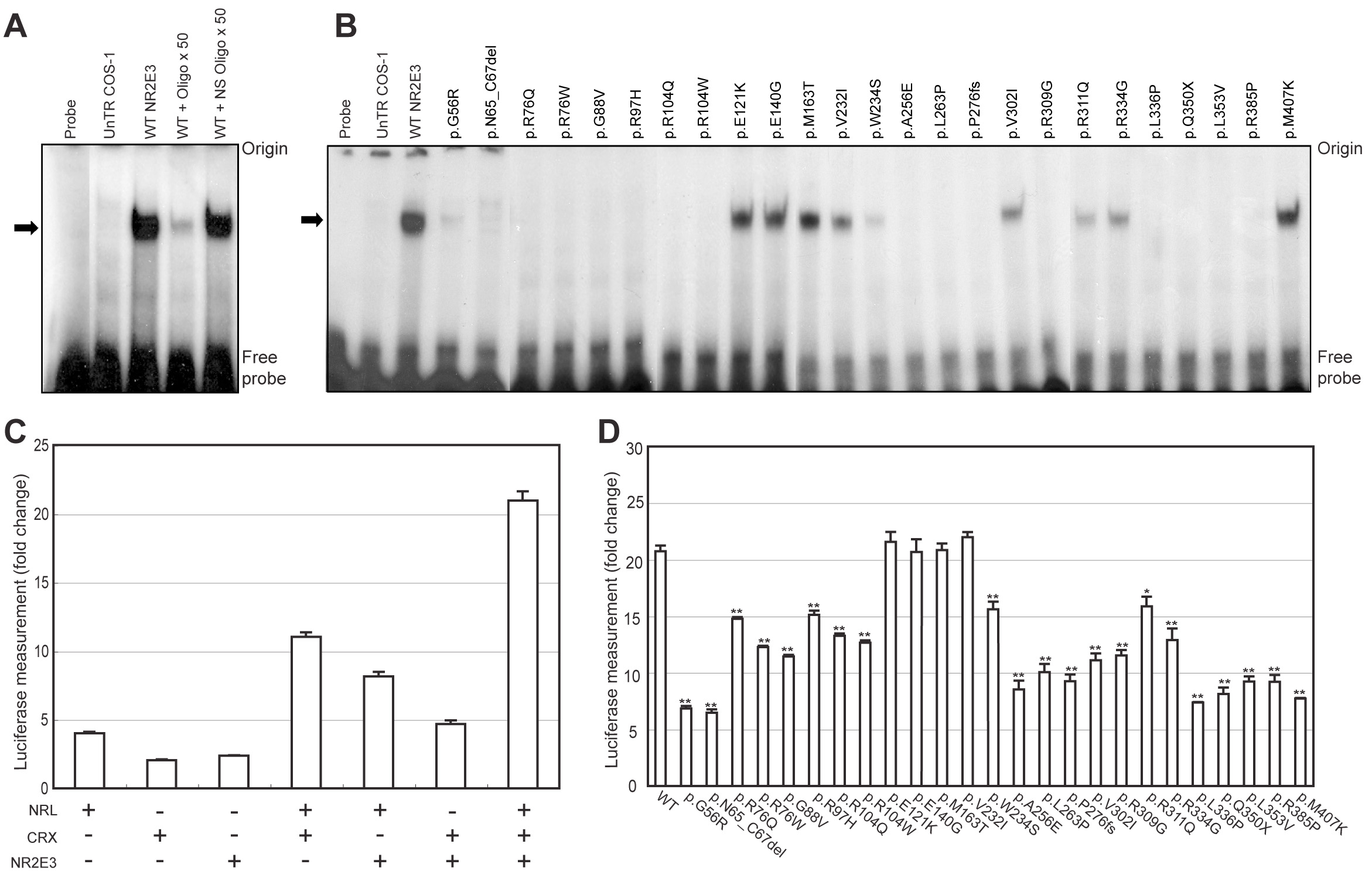
 Figure 3 of Kanda, Mol Vis 2009; 15:2174-2184.
Figure 3 of Kanda, Mol Vis 2009; 15:2174-2184.  Figure 3 of Kanda, Mol Vis 2009; 15:2174-2184.
Figure 3 of Kanda, Mol Vis 2009; 15:2174-2184. 