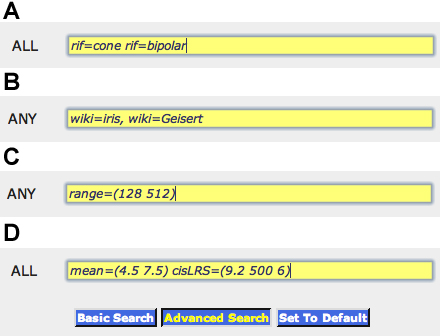
Figure 2. Advanced searching capabilities. Groups of genes, transcripts, and probe sets can be extracted from
GeneNetwork using special query commands. To review the list of commands and their syntax, click on the '
Advanced Search' button in GN, in the frame on the right side of the page. The search terms in the top panel
A “rif=cone rif=bipolar,” when placed into the ALL field of
GeneNetwork, will retrieve genes associated with cone bipolar cells, including
Atp2b1, Bsn, Gnao1, Gnb3, Gnb4, Gnb13, Grm7, Hcn1, Hcn2, Irx5, Kcnip3, and
Vsx1. This query exploits the constantly updated NCBI
GeneRIF database that is integrated into
GeneNetwork. The search terms in
B will retrieve all probe sets that have been annotated by any user in the GeneWiki with either the word “iris” or the author’s
name “geisert.” Panel
C illustrates the use of the “range” command. This command is used to find transcripts that have different levels of variation
across strains of mice. For example, the search string “
range=(128 512)” will return mRNAs assays with greater than a 128 fold and less than 512 fold difference in expression across all 103 lines
of mice. This is equivalent to a difference of 7 to 9 units (2
7 and 2
9). This search will return a list that includes
Cnga1 (cyclic nucleotide gated channel alpha 1
), Gnat1 (rod alpha transducin),
Gsn (gelsolin),
Mela (melanoma antigen),
Nrl (neural retina leucine zipper),
Pdc (phosducin),
Pde6a, Pde6b, Pde6g (three phosphodiesterases),
Rho (rhodopsin),
Rp1 (retinitis pigmentosa 1), and
Sag (S-antigen). Expression of
Sag, for example, ranges from a low of 7.9 in the
Clcn3 knockout to a high of 15.4 in PANCEVO/EiJ, the colonial mound-building mouse species. Panel
D illustrates a complex search that can be used to find probe sets with low expression but high genetic signal. This query
finds all transcripts with expression levels between 4.5 and 7.5 that are also associated with strong evidence of a linkage
peak (an LRS linkage scores >9.2 and <500) within 5 Mb of the parent gene, and where cisLRS is a shorthand to indicate that
the quantitative trait locus (QTL) is near the location of the gene and has an LRS in a defined range. The cisLRS buffer parameter
of 6 Mb in this query is equivalent to 0.5% of the mouse genome. Over 2,019 probe sets match these criteria, but there is
a limit of 2,000 probe sets to view the complete results. In comparison, a total of 6375 probe sets—15% of the content of
the array—match the query “mean=(4.5 7.5) LRS=(13.8 500).” These criteria are less restrictive and do not require transcripts
to be controlled by their own gene locus (LRS versus cisLRS). However, they do require a higher LRS threshold equivalent to
a LOD score of 3 (-logP=3, or p is approximately 0.001, where 1.0 LOD is roughly 4.6 LRS).
 Figure 2 of
Geisert, Mol Vis 2009; 15:1730-1763.
Figure 2 of
Geisert, Mol Vis 2009; 15:1730-1763.  Figure 2 of
Geisert, Mol Vis 2009; 15:1730-1763.
Figure 2 of
Geisert, Mol Vis 2009; 15:1730-1763.