Figure 1. Extracting Data from the
HEIMED. Step 1. Open the main website,
GeneNetwork. Set up the Find Records pull-down menu fields to read: Choose Species=
Mouse, Group=
BXD, Type=
Eye mRNA, Database=
Eye M430v2 (Sep08) RMA. Step 2. Make these setting your default by clicking on the '
Set to Default' button (bottom right of the window). Step 3. Enter the search term “rhodopsin” (quotes are not needed) in the ANY field
and click on the '
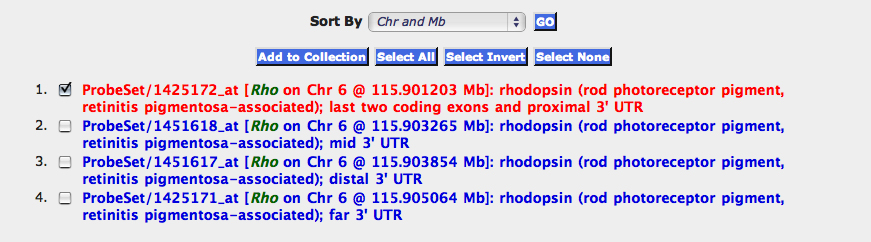
Basic Search' button. (Alternatively enter the search term “rhodopsin, rho” in the ALL field). Step 4. A Search Results window will open
with a list of seven probe sets, four of which target different parts of the rhodopsin transcript. By default the probe sets
are listed by their positional order from proximal Chr 1 to distal Chr Y. You can use the Sort By pull-down menu to reorder
probe sets by average gene expression level, symbols, or by identifier numbers. Step 5. Click anywhere on the red text to
generate a new window called the Trait Data and Analysis Form. The top of this window provides summary information on rhodopsin
and this probe set; the middle section provides Analysis Tools; and the bottom section provides a set of editable boxes that
contain the gene expression averages and error terms for all lines of mice starting with the B6D2F1 hybrids at the top and
ending with the WSB/EiJ
Mus musculus domesticus strain at the bottom (scroll to the bottom to see all of the common strains of mice).
 Figure 1 of
Geisert, Mol Vis 2009; 15:1730-1763.
Figure 1 of
Geisert, Mol Vis 2009; 15:1730-1763.  Figure 1 of
Geisert, Mol Vis 2009; 15:1730-1763.
Figure 1 of
Geisert, Mol Vis 2009; 15:1730-1763.