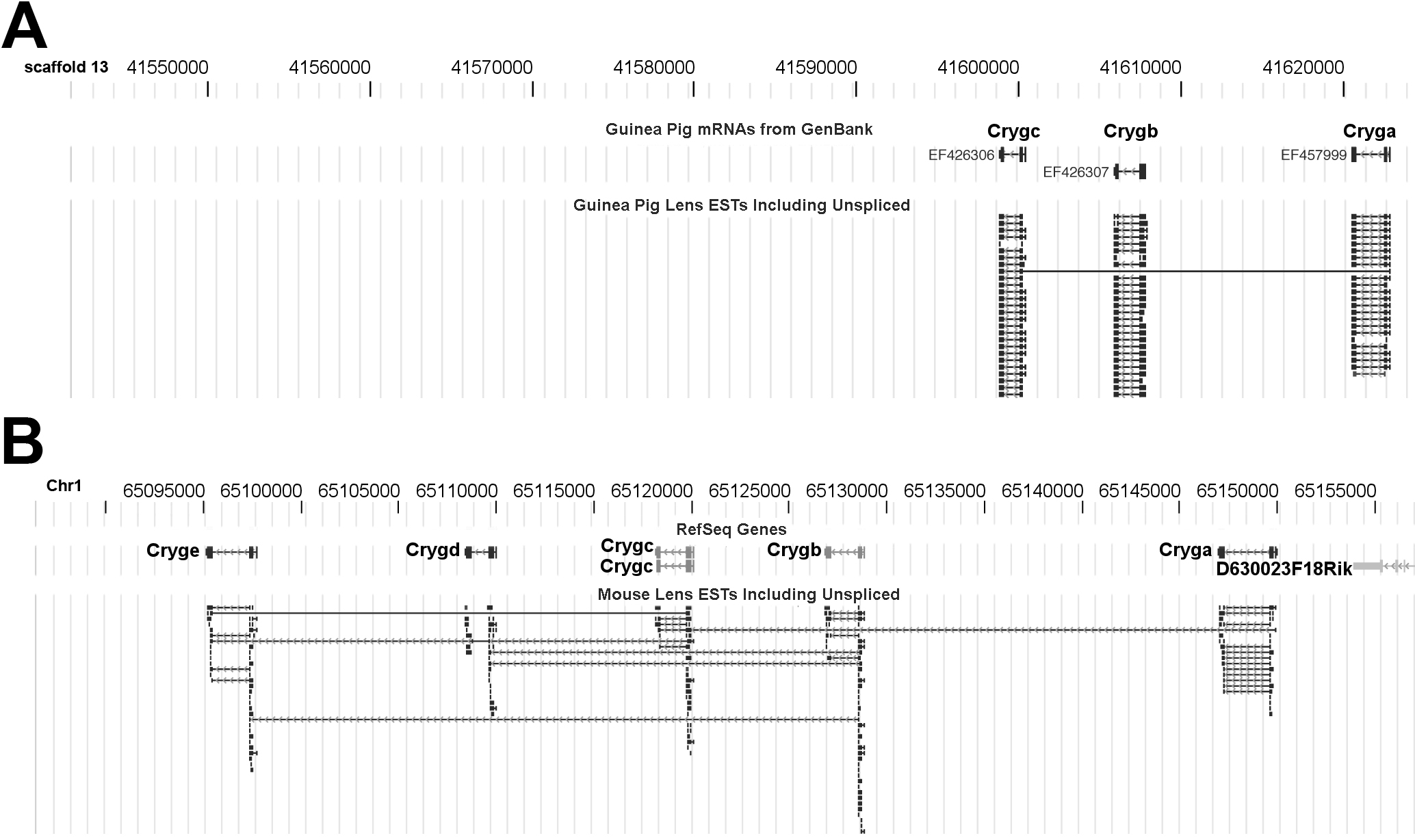
Figure 2. Absence of
Crygd, Cryde,
and
Crygf in the guinea pig genome downstream of the
Cryga-Crygc
region.
A:
EyeBrowse
view of NEIBank ESTs for guinea pig γ-crystallin
genes,
aligned to scaffold_13 of the guinea pig genome and mRNAs from
GenBank.
Cryga
(EF457999),
Crygb (EF426307) and
Crygc (EF426306), were
constructed from NEIBank EST data. There were no
BLAT
alignments to indicate the existence of guinea pig orthologs of the
Crygd,
Cryde, or
Crygf genes. In this temporary scaffold, there
were over 85,000 base pairs of gap-free DNA sequence downstream from
Crygc.
B: For comparison to the guinea pig, the mouse chromosome-1
region containing the γ-crystallin genes
Cryga to
Cryge,
aligned to RefSeq (Reference Sequence) genes. Note: arrows indicate
gene orientations, which are on the reverse strand.
 Figure 2 of Simpanya, Mol Vis 2008; 14:2413-2427.
Figure 2 of Simpanya, Mol Vis 2008; 14:2413-2427.  Figure 2 of Simpanya, Mol Vis 2008; 14:2413-2427.
Figure 2 of Simpanya, Mol Vis 2008; 14:2413-2427.