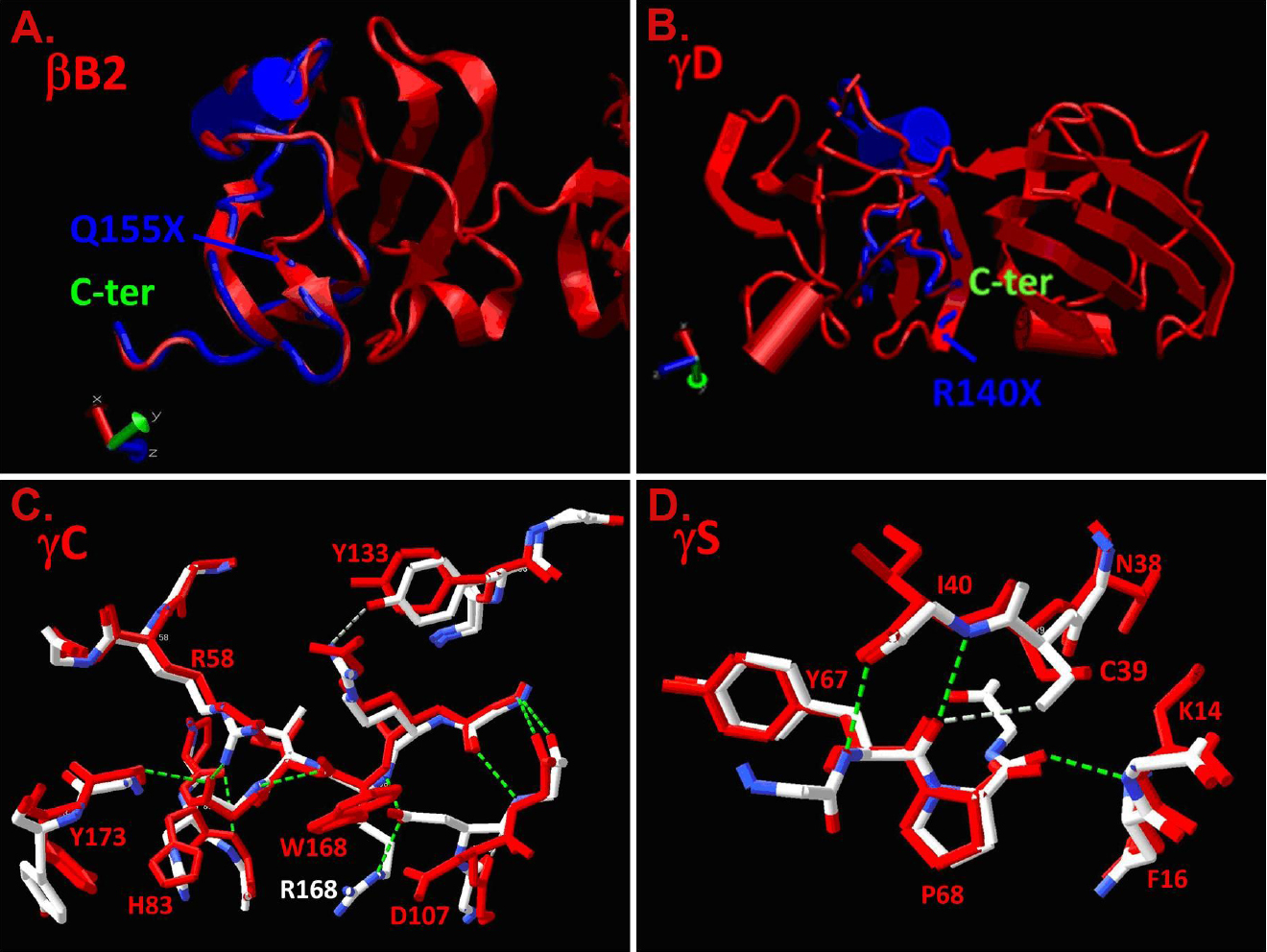
Figure 4. Molecular modeling of the
effects of mutations presented in
Table 1 on βγ-crystallin
structures. Normal structures are shown in red and the overlaid mutant
structures are shown in blue (truncations) and blue and white
(substitutions).
A: The normal βB2-crystallin structure is
shown in red, and the Q155X mutation is predicted to remove the
COOH-terminal 51 amino acids shown in the blue overlay.
B: The
normal γD-crystallin structure is shown in red and the R140X mutation
is predicted to remove the COOH-terminal 34 amino acids shown in the
blue overlay.
C: The normal γC-crystallin structure in the
vicinity of the R168R mutation is shown in white with positive charges
shown in blue and negative charges in red. The overlaid R168W mutant is
shown in red, demonstrating the substitution of the polar arginine
residue by the apolar tryptophan and consequent displacement of D107.
D:
The normal γS-crystallin structure in the vicinity of the fragments of
3D structures of mutant proteins (γC-crystallin) and S39C
(γS-crystallin) mutations are shown in red. Hydrogen-bonding patterns
of the mutant γC- and γS-crystallin structures are shown by dashed
lines (green).
![]() Figure 4 of Devi, Mol Vis 2008; 14:1157-1170.
Figure 4 of Devi, Mol Vis 2008; 14:1157-1170.