![]() Figure 3 of
Shiels, Mol Vis 2007;
13:2233-2241.
Figure 3 of
Shiels, Mol Vis 2007;
13:2233-2241.
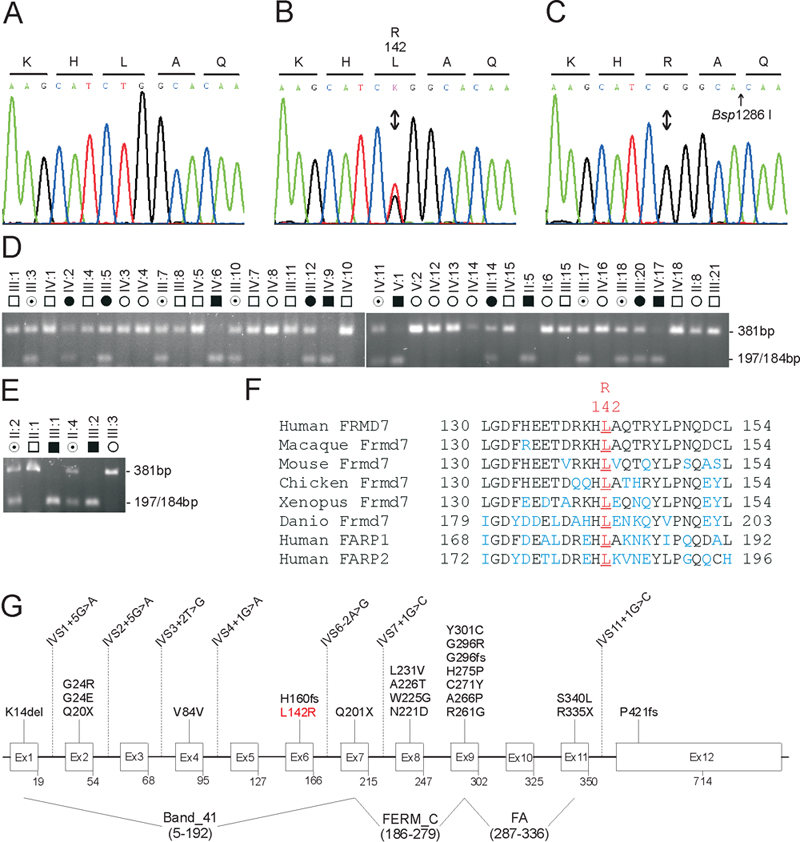
Figure 3. Mutation analysis of FRMD7
A: Sequence trace of the wild type allele, showing translation of leucine (L) at codon 142 (CTG). B: Sequence trace of the heterozygous mutant allele, showing the c.425T>G transversion (denoted as K by the International Union of Pure and Applied Chemistry [IUPAC] code) that is predicted to result in the missense substitution of arginine (R) for leucine at codon 142 (p.L142R). C: Sequence trace of the hemizygous mutant allele showing translation of arginine (R) at codon 142 (CGG). D-E: Restriction-fragment-length analysis showing gain of a Bsp1286I site (5'-GGGCA/C) that cosegregates only with affected and carrier individuals hemizygous or heterozygous for the c.425T>G transversion (197/184 bp) in family W (D) and family M (E). F: Amino acid sequence alignment of human FRMD7 and orthologs from other species, showing phylogenetic conservation of p.L142. G: Exon organization and mutation profile of FRMD7. Codons are numbered below each exon. So far, more than 30 mutations have been reported [20-24]. Note the synonymous p.V84V polymorphism (c.252G>A) creates a cryptic splice acceptor-site in exon 4 resulting in missplicing [20].