![]() Figure 3 of
Yip, Mol Vis 2007;
13:2183-2193.
Figure 3 of
Yip, Mol Vis 2007;
13:2183-2193.
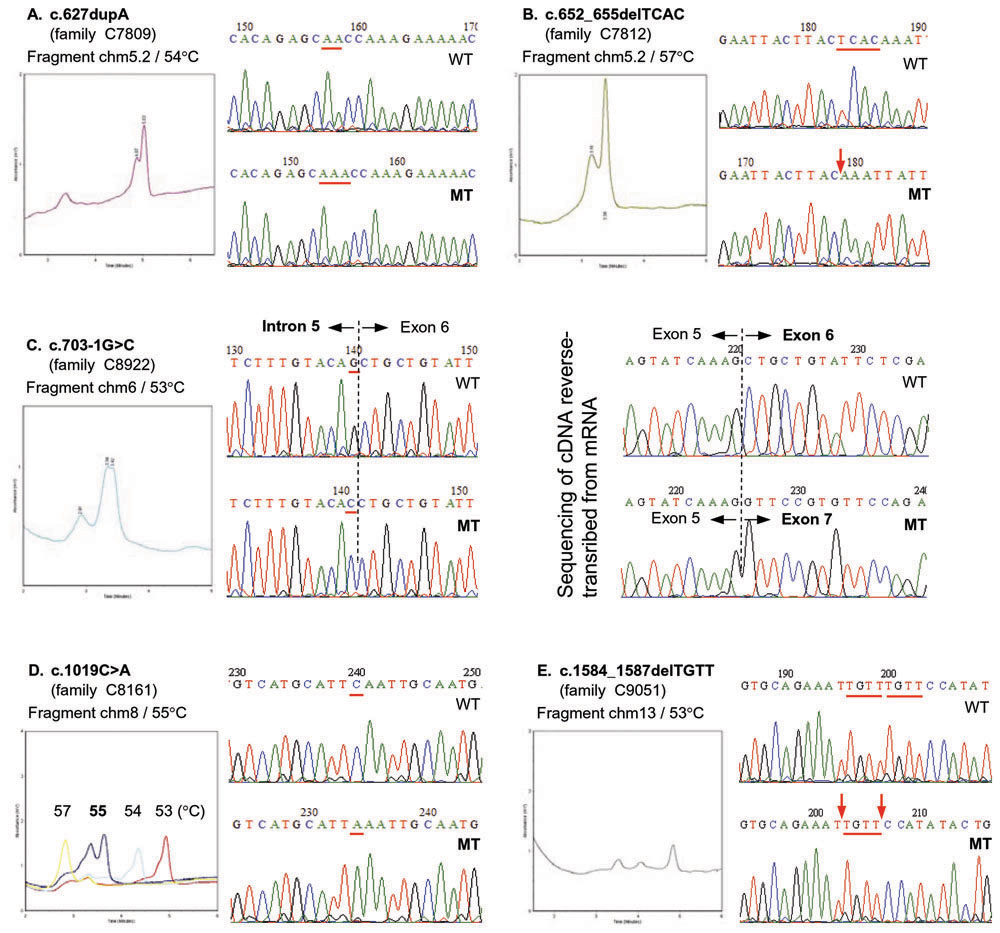
Figure 3. Mutation screening by denaturing high performance liquid chromatography and characterization by DNA sequencing
Each part (A, B, C, D, or E) is a mutation identified in a particular family. It consists of a chromatogram on the left, and two DNA sequencing traces on the right with wildtype sequence (WT) on top of the mutant sequence (MT). For the chromatogram, the PCR fragment of interest and the column temperature for analysis are indicated. The elution time is indicated in the x-axis and the absorbance at 260 nm in the y-axis. For part D, the chromatograms for four column temperatures are overlaid to illustrate the use of multiple column temperatures during screening; only 55 °C gave a distinctive two-peak elution profile. For DNA sequencing traces, the bases are underlined if they were affected (for parts B to D) or if they formed the repeating units that were inserted or deleted (parts A and E). B: The downward arrow indicates the position where four bases were deleted. C: The sequencing results for cDNA are also included to show that the whole exon 6 is deleted as a result of the splicing mutation c.703-1G>C. E: The two downward arrows indicate that deletion of the first or the second TGTT unit gave the same result.