![]() Figure 1 of
Yuan, Mol Vis 2007;
13:2083-2095.
Figure 1 of
Yuan, Mol Vis 2007;
13:2083-2095.
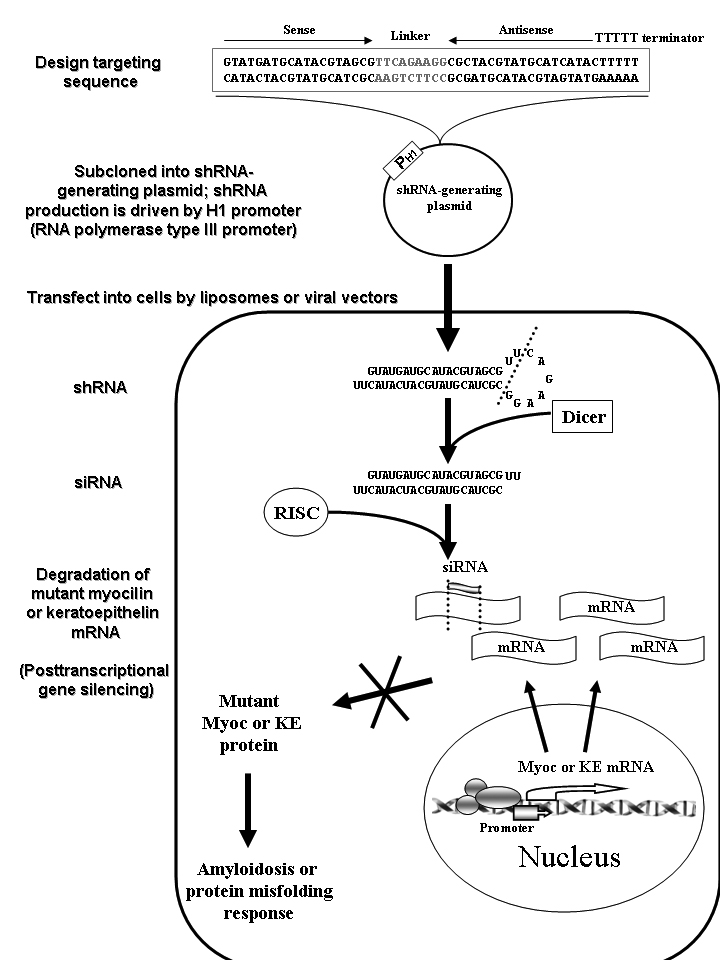
Figure 1. Gene silencing by short hairpin RNA
RNA interference (RNAi)-mediated gene silencing is a posttranscriptional mechanism targeting a specific mRNA in which a short double-stranded RNA of 21-23 nucleotide pairs (known as "small interfering RNA", siRNA) induces a sequence-specific knock-down of its complementary gene. To prevent degradation of RNA fragments and to achieve a stable expression of siRNA in cultured cells, self-looped short hairpin RNAs (shRNAs) as siRNA precursors can be designed by linking the sense and antisense strands of siRNA with an oligonucleotide linker (spacer) and a poly-T as a terminator. The sequences can then be subcloned into a plasmid, containing RNA polymerase III promoters, such as human H1-RNA promoter to generate shRNAs. These plasmids can transfect target cells via liposomes or viral vectors. Once inside the cell, the shRNAs generated by the plasmids are cleaved by a Dicer (an RNase) to form siRNAs. The double stranded siRNA is unwound to form a single-stranded ribonucleoprotein complex known as RNA-induced silencing complex (RISC), which mediates a sequence-specific degradation of the mRNAs involved in coding a target protein. After pairing with a siRNA strand, the target mRNA is cleaved and further degraded, leading to an interruption in synthesis of the disease-causing protein such as myocilin or keratoepithelin (KE). The RISC complex is naturally stable thus enabling siRNAs to interact consecutively with multiple mRNAs with a potent suppression of protein synthesis. With suppression of mutant KE and myocilin, the amyloidogenic response from aggregations of abnormal KE and the cytotoxic response of trabecular meshwork caused by misfolded myocilin can be mitigated, respectively.