![]() Figure 5 of
Li, Mol Vis 2007;
13:1758-1768.
Figure 5 of
Li, Mol Vis 2007;
13:1758-1768.
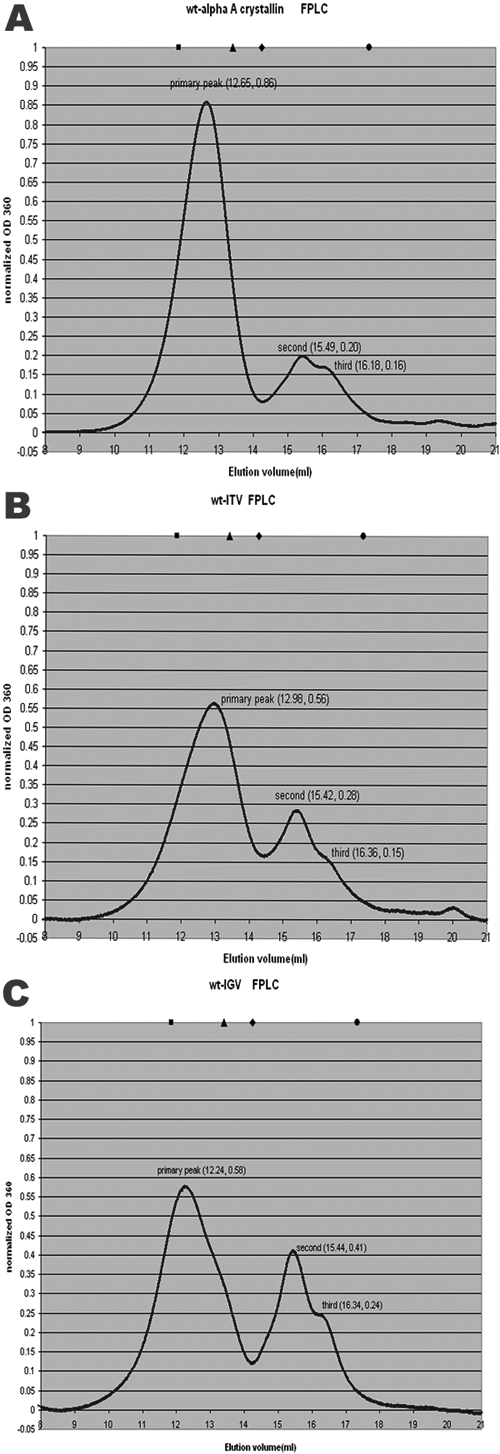
Figure 5. Size exclusion chromatograms for wild-type αA and mutants
FPLC was performed using a Superose 6 column calibrated with molecular weight standards as described in Methods and run with 0.1 M NaCl Tris buffer, pH 8.0. A-C are the FPLC profiles of wt-αA-crystallin, wt-ITV, and wt-IGV, respectively. The y-axis value, OD360, was normalized by Origin 6.1 so that the area under the curve of each mutant was the same. The positions of proteins used for standard molecular masses are also indicated at the top of the diagram. The solid black square indicates thyroglobulin (669 kDa), the triangle indicates apoferritin (443 kDa), the diamond indicates β-amylase (200 kDa), and the dot indicates alcohol dehydrogenease (150 kDa).