![]() Figure 1 of
Qin, Mol Vis 2006;
12:485-491.
Figure 1 of
Qin, Mol Vis 2006;
12:485-491.
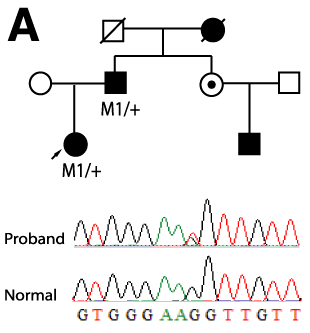
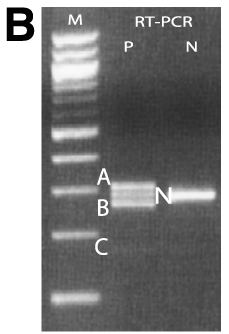
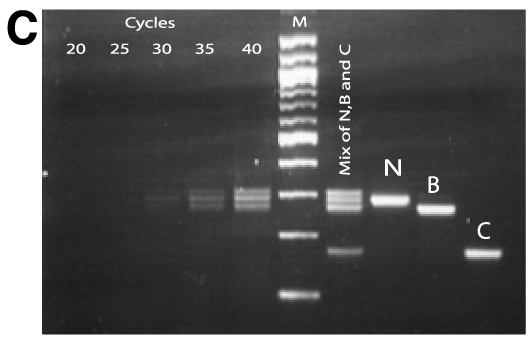
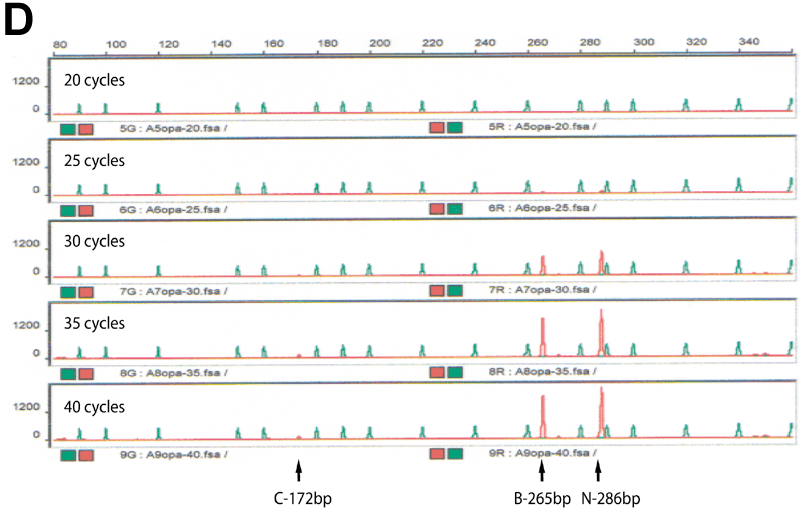
Figure 1. Identification of a novel splicing mutation and characterization of their RNA in a Family 1
A: Top panel is the pedigree of the Family 1. Arrow indicates the proband and the filled solid symbols show the affected individuals. Genotypes of individuals from whom sequence data were obtained are presented. Plus signs indicate the wild type and M1 indicates the splicing mutation of c.871-1G>T. The bottom panel represents the sequence chromatograms of the proband (upper) and that of normal individual (lower) with an arrow indicating the nucleotide change. B: Ethidium bromide-agarose gel loaded RT-PCR products generated from leukocytes of total RNA of proband and a normal sample along with 100 bp size marker. C: Ethidium bromide-agarose gel loaded RT-PCR products of the proband. On the left of the size marker (100 bp ladder) were RT-PCR products from different numbers of PCR amplification cycle. On the right of the size marker (100 bp ladder) was the mixture of band B, C, and N along with gel purified individual DNA samples of B, C and N. D: Denaturing capillary electrophoresis profile of RT-PCR products of the proband after different numbers of PCR amplification cycle. E: DNA and amino acid sequences alignment for mutant and wild type transcripts. A cryptic acceptor site is red color.
E:
114 bp deletion attgcagaaagatgacaaaggcattcatcatagaaagcttaagaaatctttgattgacatgtattctgaagttcttgatgttctctctga 90
L Q K D D K G I H H R K L K K S L I D M Y S E V L D V L S D
21 bp deletion attgcagaaagatgacaaaggcattcatcatagaaagcttaagaaatctttgattgacatgtattctgaagttcttgatgttctctctga 90
L Q K D D K G I H H R K L K K S L I D M Y S E V L D V L S D
OPA1-wildtype attgcagaaagatgacaaaggcattcatcatagaaagcttaagaaatctttgattgacatgtattctgaagttcttgatgttctctctga 90
L Q K D D K G I H H R K L K K S L I D M Y S E V L D V L S D
114 bp deletion ttatgatgccagttataatacgcaagatcatctgccacgg-------------------------------------------------- 130
Y D A S Y N T Q D H L P R 38aa deletion of entire exon 9
21 bp deletion ttatgatgccagttataatacgcaagatcatctgccacgg---------------------agtgctggaaagactagtgtgttggaaat 159
Y D A S Y N T Q D H L P R 7aa deletion S A G K T S V L E M
OPA1-wildtype ttatgatgccagttataatacgcaagatcatctgccacgggttgttgtggttggagatcagagtgctggaaagactagtgtgttggaaat 180
Y D A S Y N T Q D H L P R V V V V G D Q S A G K T S V L E M
114 bp deletion ----------------------------------------------------------------gtgactctgagtgaaggtcctcacca 156
V T L S E G P H H
21 bp deletion gattgcccaagctcgaatattcccaagaggatctggggagatgatgacacgttctccagttaaggtgactctgagtgaaggtcctcacca 249
I A Q A R I F P R G S G E M M T R S P V K V T L S E G P H H
OPA1-wildtype gattgcccaagctcgaatattcccaagaggatctggggagatgatgacacgttctccagttaaggtgactctgagtgaaggtcctcacca 270
I A Q A R I F P R G S G E M M T R S P V K V T L S E G P H H
114 bp deletion tgtggccctatttcca 172
V A L F P
21 bp deletion tgtggccctatttcca 265
V A L F P
OPA1-wildtype tgtggccctatttcca 286
V A L F P
|