![]() Figure 1 of
Devi, Mol Vis 2006;
12:190-195.
Figure 1 of
Devi, Mol Vis 2006;
12:190-195.
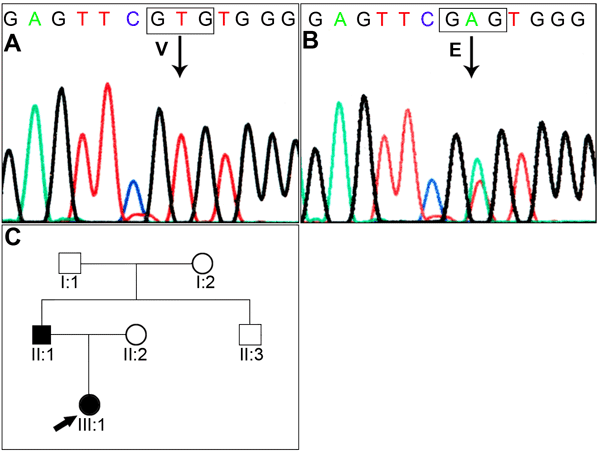
Figure 1. Analysis of family 1
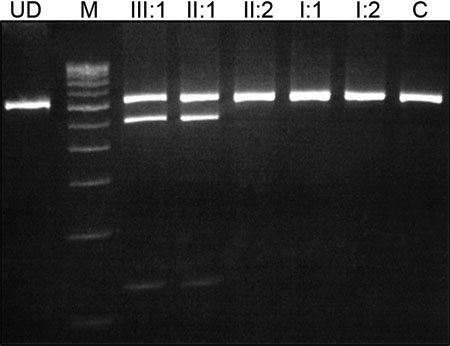
A: The genomic sequence of the wild type showing valine at codon 44. B: The mutant allele showing a heterozygous T>A transversion that replaces Val by Glu. C: Pedigree of family 1. D: PCR-RFLP analysis showing a gain of a TaqI site that cosegregates with the affected individuals. The unaffected individuals display a 648 bp band while the affected individuals display a 648 bp, 519 bp, and 129 bp bands. In the image, M indicates the 100 bp DNA ladder, C indicates control, and UD indicates undigested PCR product. E: A multiple alignment of amino acid sequence of GJA8 with different species and five other connexins from human. The amino acid marked in red indicates a neutral amino acid (valine, alanine, or serine) at codon 44.
D:
E:
CXA8_HUMAN 11 LEEVNEHSTVIGRVWLTVLFIFRILILGTAAEFVWGDEQS 50 CXA8_MOUSE 11 LEEVNEHSTVIGRVWLTVLFIFRILILGTAAEFVWGDEQS 50 CXA8_SHEEP 11 LEEVNEHSTVIGRVWLTVLFIFRILILGTAAEFVWGDEQS 50 CXA8_CHICK 11 LEQVNEQSTVIGRVWLTVLFIFRILILGTAAELVWGDEQS 50 CXA1_HUMAN 11 LDKVQAYSTAGGKVWLSVLFIFRILLLGTAVESAWGDEQS 50 CXA3_HUMAN 11 LENAQEHSTVIGKVWLTVLFIFRILVLGAAAEDVWGDEQS 50 CXA4_HUMAN 11 LDQVQEHSTVVGKIWLTVLFIFRILILGLAGESVWGDEQS 50 CXA5_HUMAN 11 LEEVHKHSTVVGKVWLTVLFIFRMLVLGTAAESSWGDEQA 50 CXA10_HUMAN 11 LEEVHIHSTMIGKIWLTILFIFRMLVLGVAAEDVWNDEQS 50 |