![]() Figure 6 of
Nickerson, Mol Vis 2006;
12:1565-1585.
Figure 6 of
Nickerson, Mol Vis 2006;
12:1565-1585.
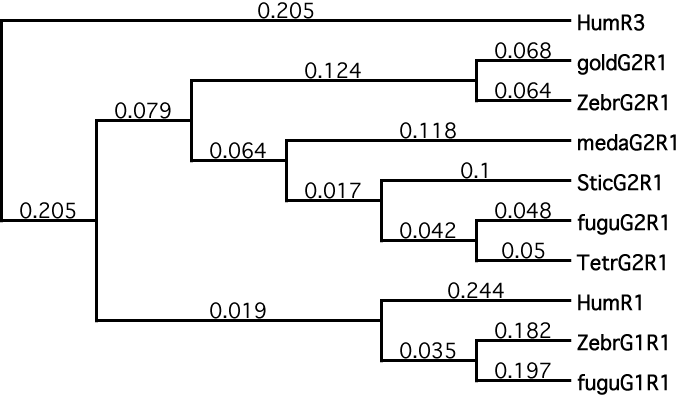
Figure 6. Phylogenetic analysis of Repeat 1 from IRBP
Evolutionary relationships among fish and human Repeat 1 orthologs and paralogs are illustrated. MacVector version 9.0 was used to build a cladogram of the Repeat 1-like amino acid sequences of the teleost fish and the human Repeat 1. Human Repeat 3 was used as an outgroup for the purpose of rooting this tree. The illustrated tree was built using the neighbor-joining method. Whether using the UPGMA or neighbor-joining method, with uncorrected or Poisson-corrected distances, random or systematic tie-breaking for gaps, in MacVector version 9.0, all the phylogenetic reconstructions gave the same overall structure. A consensus tree from 1000 replications of bootstrapping was calculated, and it was identical to the structure of the best tree, which is illustrated in this image. Among 1000 replications of resampling, all seven relevant nodes occurred frequently, with four appearing in 100% of trees, two nodes occurring in 96% of trees, and one node occurring in 79% of trees. All the nodes are resolved in the illustrated tree. The results suggest a paralogous relationship between the Repeat 1 sequences from Genes 1 and 2 in zebrafish and fugu. The closer relationship of human Repeat 1 to the fugu and zebrafish Gene 1 orthologs than to Gene 2 and the illustrated divergence at the leftmost internal node of the paralogs on this cladogram suggests that the paralogous Genes 1 and 2 arose early or at the origin of the teleosts. This tree is consistent with the already known divergence radiation of the teleost fish about 350 Mya.