![]() Figure 1 of
Okamoto, Mol Vis 2006;
12:156-158.
Figure 1 of
Okamoto, Mol Vis 2006;
12:156-158.
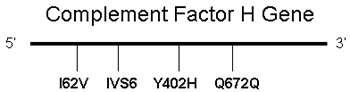
Figure 1. Association analysis of Complement Factor H haplotypes
A: Four sets of single nucleotide polymorphisms (SNPs; I62V, IVS6, Y402H, Q672Q) were selected to distinguish all five haplotypes reported [6]. B: All haplotypes with a frequency >3% are displayed. The estimated frequencies for Japanese (JP) and American (US) cases and controls in these risk and protective haplotypes are shown (JP/US). The red "C" represents the nucleotide responsible for the Y402H polymorphism. N/A means "not available."
A:
B:
Estimated frequencies
------------------------- Risk (R) or
Cases Controls protective (P)
----------- ----------- likelihood
Haplotypes p JP US JP US (fold JP/US)
-------------- ----- ---- ---- ---- ---- --------------
H1: G T T G 1.000 0.38 N/A 0.38 N/A 1.0/-
H2: A T T A 0.001 0.18 0.12 0.35 0.21 (P) 1.6/1.7
H3: G T T A 0.028 0.19 N/A 0.10 N/A (R) 1.9/-
H4: A T T C 0.004 0.15 N/A 0.06 N/A (R) 2.5/-
H5: G T C A 0.802 0.04 0.50 0.04 0.29 (R) 1.0/1.7
|