![]() Figure 1 of
Vieira, Mol Vis 2006;
12:1448-1460.
Figure 1 of
Vieira, Mol Vis 2006;
12:1448-1460.
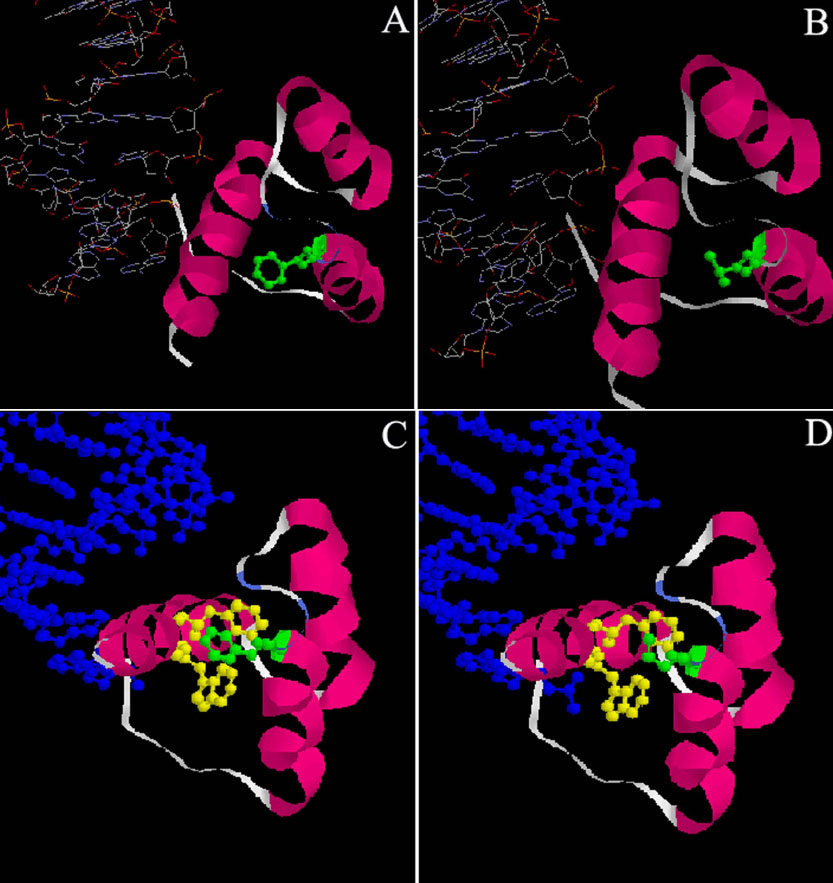
Figure 1. Crystallographic views of the phenylalanine 58 to leucine (F58L) mutation in PITX2 protein using Rasmol software
The PITX2 crystallographic views were obtained and derived from the structural model obtained by X-ray diffraction of the Drosophila engrailed homeodomain protein (pdb: 1HDD). The general view (A) localizes precisely the residue Phe20 (position in the homeodomain). Phe20 is situated at the end of the helix 2, between helix 1 and helix 3 (pink), which recognizes and interacts with DNA (blue).The crystallographic view (B) shows that the side chain (green) of the residue Leu is shorter and has different sterical properties than the residue Phe. As a consequence, the distances between atoms should be longer in the case of the mutant protein F20L. However, the hydrophobic nature of the side chain is conserved. The local views of the environment of the residue Phe20 (C and D) show the side chains of the close residues Phe49 and Trp48 in helix 3 (yellow). This environment allowed a network of hydrophobic interactions with Phe20 (green), which stabilize the homeodomain of engrailed protein and also of PITX2 protein. The mutation F58L does not change the nature of the potential interactions, but the shorter side chain will either cause a structural rearrangement or will increase the distances between the atoms, probably affecting the conformation of the homeodomain.