![]() Figure 3 of
Singh, Mol Vis 2006;
12:1372-1379.
Figure 3 of
Singh, Mol Vis 2006;
12:1372-1379.
Figure 3.
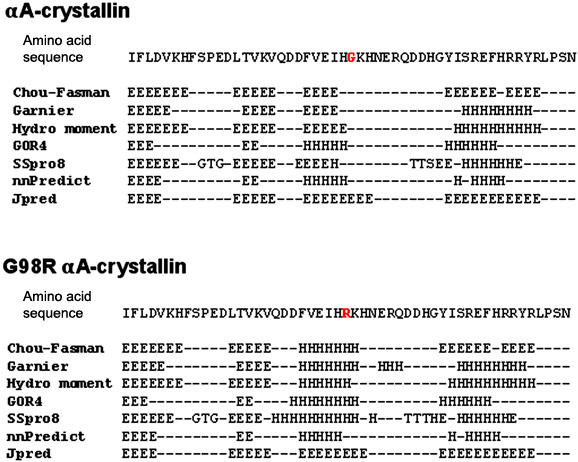
Prediction of the secondary structural propensity of the sequence of the wild type and G98R αA-crystallin by various programs using different algorithms. The full-length sequences were subjected to predictions using Chou-Fasman, Garnier, hydrophobic moment (using software PepTool Lite 1.1), GOR4, SSpro8, nnPredict, Jpred programmes. Secondary structural propensities of the sequence from residue 73 to 123 are shown. Some of the programs predict a change in propensity from strand (E) in the wild type to helical (H) in the mutant in the sequence surrounding the mutated amino acid (shown in red). H, helix; E, extended strand; T, turn; G, 310 helix; S, bend and Dashed line, random coil.