![]() Figure 1 of
Sun, Mol Vis 2006;
12:1364-1371.
Figure 1 of
Sun, Mol Vis 2006;
12:1364-1371.
Figure 1.
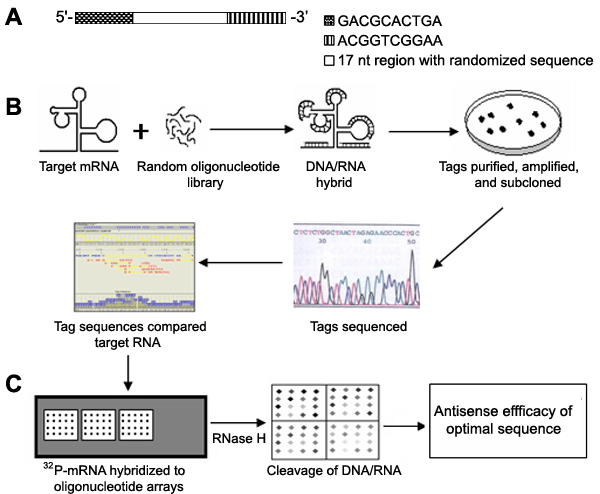
Schematic diagram of full length gene targeting (FLGT). A: An oligonucleotides library with 17 nt fully randomized sequence nested between two fixed sequences for binding to PCR primer. B: mRNA was synthesized by in vitro transcription and hybridized with fully oligonucleotides library. The oligonucleotide tags specifically bound to the mRNA were PCR amplified, cloned into vectors, sequenced and aligned to target gene. The selection of accessible sites and design of antisense oligonucleotides (ASOs) were accomplished by TargetFinder software. C: Oligonucleotide probes were designed and constructed microarrays. Radiolabeled transcripts (cRNA) hybridized to microarrays. The hybridization signal reflects the degree of binding affinity between cRNA and oligonucleotides. Subsequently, the heteroduplexes were followed by RNase H reaction for various times. According to the residuary signal intensity with time-course, RNase H cleavage ability for oligonucleotide-RNA duplexes was estimated by T1/2 (the time of 50% heteroduplex cleavaged by RNase H).