![]() Figure 3 of
Hansen, Mol Vis 2006;
12:1033-1039.
Figure 3 of
Hansen, Mol Vis 2006;
12:1033-1039.
Figure 3.
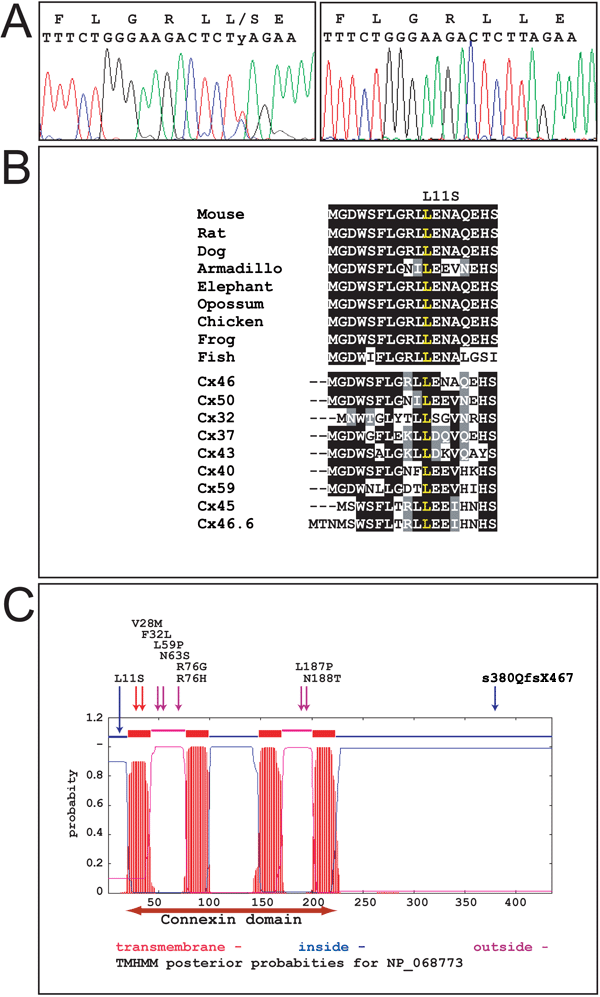
Protein sequence and structure of various connexins. A: Chromatogram of the GJA3 DNA sequence from individual II:8 carrying the mutations (left) and an individual having wild type alleles. A double peak at position 32 represents the T>C transition and a shift of leucine to serine in the signal peptide. B: ClustalW aligments of the first 18 amino acids of the signal peptide of Cx46 in various animals. The sequences are: mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis familiaris) armadillo (Armadillo officinalis), elephant (Loxodonta africana), opossum (Monodelphis domestica), chicken (Gallus gallus), frog (X_tropicalis), and fish (Tetraodon nigroviridis). The DNA sequences used for translation are from the latest assemblies from GenBank and [23]. Below the aligned sequences for the different species are the aligned signal peptide sequences for 9 different human connexin proteins. Cx46 and Cx50 are the only lens-specific expressed proteins. The conserved leucine at position 11 is marked in yellow. The human sequences are derived from the following GenBank accession numbers: Cx46, NP_068773; Cx50, NP_005258; Cx32, NP_000157; Cx37, NP_002051; Cx43, NP_000156; Cx40, NP_005257; Cx59, NP_110399; Cx45, NP_005488; and Cx46.6, NP_065168 [23]. C: Prediction of the transmembrane helices in the Cx46 protein shown in graphics. The location and orientation of domains in the primary protein structure are shown on the X-axis and the probability for forming αhelices is shown on the Y-axis [10]. Cytoplasmatic, transmembrane, or extracellular regions are shown in color on top of the plot. A horizontal red two-headed arrow below the plot indicates the conserved connexin-domain. All known human Cx46 mutations [13-19] are denoted above the plot. Transmembrane mutationsare identified by red arrows, mutations in the cytoplasmatic regions are identified by blue arrows, and mutations in the extracellular domains are identified by violet arrows.