![]() Figure 1 of
Hanna, Mol Vis 2006;
12:961-976.
Figure 1 of
Hanna, Mol Vis 2006;
12:961-976.
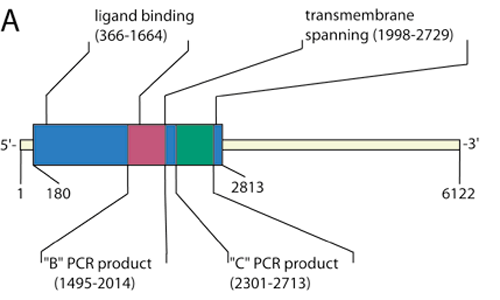
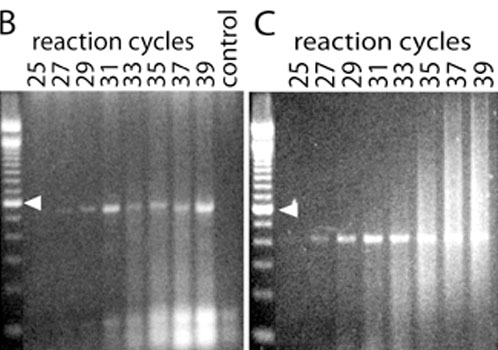
Figure 1. Sequence and expression of mGluR6 mRNA in primate retina
A: Schematic of gene encoding mGluR6 (6122 base pairs, Genbank NM_000843), indicating the coding region (blue area) and ligand-binding and transmembrane-spanning regions. Red and green areas represents highly specific regions targeted for PCR probing. B: Gene-specific PCR to detect mGluR6 within retinal cDNA library created using TPEA RT-PCR. The decrease in PCR product at 33 to 35 cycles is likely due to increased primer dimerization, indicated by the smears at the bottom of the gel. No mGluR6 mRNA is detected when reverse transcriptase is absent (control lane). Gene-specific primers were designed to produce a PCR product of 520 base pairs: 5'-GGA TGC TTC TGC AGT ACA TTC GAG-3' (sense) and 5'-TTG TTG TGT CGC ATG AAG GTG GCC-3' (antisense). C: Second gel demonstrating linear range for detection of mGluR6 using primers designed against a smaller segment of the coding region. Gene-specific primers were designed to produce a PCR product of 413 base pairs: 5'-CAG GTG GTG GGG ATG ATA GCA TG-3' (sense) and 5'-AAG AGG ATG ACG TAG GTT TTG GG-3' (antisense). In this case, there is less dimerization and the PCR product increases as expected with increasing number of cycles. Arrowhead again indicates 600 base pairs.