![]() Figure 3 of
Jiang, Mol Vis 2005;
11:143-151.
Figure 3 of
Jiang, Mol Vis 2005;
11:143-151.
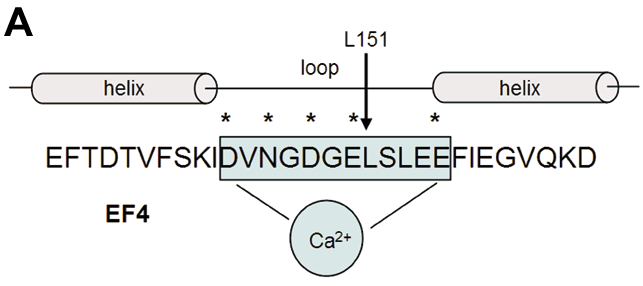
Figure 3. The EF4 Ca2+ binding site and sequence alignment of vertebrate GCAPs
A: The Ca2+ binding loop consisting of 12 amino acids is flanked by two a-helical regions. Residues in the loop that coordinate the Ca2+ ion are marked with an asterisk. Although the 151 position amino acid (arrow) does not directly coordinate the Ca2+ ion, it is highly conserved throughout the GCAPs (leucine or isoleucine) suggesting it is essential for structure and function of this loop. B: The known vertebrate guanylate cyclase activating proteins (vertebrate GCAP1-8) have been aligned (only the C-terminal halves containing EF3 and EF 4 are shown). Residues conserved in more than 50% of the sequences shown are red. Conservative substitutions are blue (for origin of sequences and accession numbers, see Methods).
B:
L151F
I143NT | E155G
Y99C | | |
| EF3 | EF4 | |
|------------ |------------
hGCAP1 94 WYFKLYDVDGNGCIDRDELLTIIQAIRAINPCSDTTMTAE---------EFTDTVFSKIDVNGDGELSLEEFIEGVQKDQMLLDTLTRSLDLTRIVRRLQNGEQDEEGADEAAEAAG
bGCAP1 94 WYFKLYDVDGNGCIIRDELLTIIRAIRAINPCSDSTMTAE---------EFTDTVFSKIDVNGDGELSLEEFMEGVQKDQMLLDTLTRSLDLTRIVRRLQNGEQDEEGASGRETEAAEADG
mGCAP1 94 WYFKLYDVDGNGCIDRDELLTIIRAIRTINPWSDSSMSAE---------EFTDTVFAKIDINGDGELSLEEFMEGVQKDQMLLDTLTRSLDLTGIVRRLQNGEHEEAGTSDLAAEAAG
ratGCAP1 94 WYFKLYDVDGNGCIDRDELLTIIRAIRTINPWSDSSMSAE---------EFTDTVFAKIDINGDGELSLEEFMEGVQKDQMLLDTLTRSLDLTRIVRRLQNGEQEEAGAGDLAAEAAG
fGCAP1 93 WYFKLYDVDGNGCIDRGELLNIIKAIRAINRCN-DEMTAE---------EFTDMVFDKIDINGDGELSLEEFIEGVQKDELLLEVLTRSLDLKHIVYMIQNDGKRMEISERPRQEITTGNSLP
cGCAP1 93 WYFKLYDVDGNGCIDRGELLNIIKAIRAINRCN-EAMTAE---------EFTNMVFDKIDINGDGELSLEEFMEGVQKDEVLLDILTRSLDLTHIVKLIQNDGKNPHAPEEAEEAAQ
oryGCAP1 93 WYFKLYDVDGNGCIDRHELLNIIKAIRAINGNENQEMTAE---------EFTNNVFDRIDVNGDGELSLEEFVEGARSDEDFMEVMMKSLDLRHIVAMIHNRRHSV
fuguGCAP1 93 WYFKLYDVDGNGCIDRHELLNIIKAIRAINGNENHETTAE---------DFTNSVFDRIDINGDGELSLEEFVAGARSDDVFMEVMIKSLDLTHIVAMIHNRRHSV
zGCAP1 93 WYFRLYDVDGNGCIDRYELLNIIKAIRAINGSETQESSAE---------EFTNRVFERIDINGDGELSLDEFVAGARSDEEFMEAMMKSLDLTHIVAMIHNRRHSV
bGCAP2 99 WTFKIYDKDRNGCIDRQELLDIVESIYKLKKACSVEVEAEQQGKLLTPEEVVDRIFLLVDENGDGQLSLNEFVEGARRDKWVMKMLQ--MDLNPSSWISQQRRKSAMF
hGCAP2 96 WTFKIYDKDGNGCIDRLELLNIVEGIYQLKKACRRELQTEQD-QLLTPEEVVDRIFLLVDENGDGQLSLNEFVEGARRDKWVMKMLQ--MDMNPSSWLAQQRRKSAMF
mGCAP2 96 WTFKIYDKDRNGCIDRLELLDIVEAIYKLKKACRAELDLEHQGQLLTPEEVVDRIFLLVDENGDGQLSLTEFIEGARRDKWVMKMLQ--MDINPGGWITQQRRRSAMF
fuguGCAP2 95 WTFKMYDKDGSGCIDKTELLEIVESIYRLKKACHGELDEDCT--LLTPDQVVDRIFELVDENGDGELSLDEFIDGARRDKWVMKMLQ--MDVNPGDWLND-RRRSADF
oryGCAP2 95 WTFKMYDKDGSGCIDKTELLEIVESIYRLKKACHGELDEECN--LLTPDQVVDRIFELVDENGDGELSLDEFIDGARRDKWVMKMLQ--MDVNPGDWINE-RRCSEDF
zGCAP2 94 WTFKMYDKDGSGCIDKTELKEIVESIYRLKKACHGELDAECN--LLTPDQVVARIFELVDENGDGELSLDEFIAGARRDKWVMKMLQ--MDVNPGDWINEQRRRSANF
fGCAP2 94 WTFKVYDRDGNGCIDKTELLEIVESIYNLKKVCRQGQDDRIP--LLSPEQVVDRIFQLVDENGDGQLSLDEFIDGARKDKWVMKMLQ--MDVSPGSWINEQRRKSALF
cGCAP2 95 WTFKVYDKDGNGCIDKPELLEIVESIYKLKKVCRSEVEERTP--LLTPEEVVDRIFQLVDENGDGQLSLDEFIDGARKDKWVMKMLQ--MDVNPGGWISEQRRKSALF
siluGCAP2 92 WSFKIYDKDGNGCVDKRELKEIIQSIYSIKRGWRR--DQEAQ--LMSPEEICERIFQIVDENGDGQLSLQEFVEGAXKDTWVLKMLQ--LDTNPCNWVMEQRRKSALF
fuguGCAP6 93 WSFKVYDRDNNGFVDRTELRSIIDSIHRIKK------DSGSQ---LTVDEVVDRIFQAADSDGDGYISAEEFIRGAQQDPWLLNILK--LDMNPAGWVMEQRRKSAHF
fuguGCAP7 92 WSFKMYDKDGNGKLDRQEVKRLIKIIHKIKLQRSDINMTP--------SEICDRIFELVDNNNDGQISLSEFMEGAQKDEWVMNLLK--LDVNATSWVLHNCEKLP
fuguGCAP8 94 WSFKVYDRDGNGCLDKQEVRHIVKIIHKIK-KKSDMSVTEKI------EDVCDRIFELVDKNKDSQISLEEFIEGAEKDPWLMNQLR--LDIGPCEWFIEQREKKL
fuguGCAP5 92 WYFKLYDIDGSGCIDRDELLLIIKSIRAINGIP-SEMSAE---------EFTNMVFDKIDINGDGELSYEEFIEGIQNDETLLKMLTESLDLTHIMQKIEGQMNASSSD
zGCAP5 93 WYFKLFDMDGSGCIDKDELLLIFKAVQAINGAE-PEISAE---------DLADMVFNKIDVNGDGELSLEEFMEGISADEKISEMLTQSLDLTRIVSNIYNDSYIEQEAEIIEDQA
zGCAP3 92 WYFKLFDQDGNGKIDRDEMETIFKAIQDITRSY--EIPPD---------DIVSLIYERIDVNNEGELTLEEFITGAKEHPDIMEMLTKMMDLTHVLEIIVNGQKKKKE
fuguGCAP3 92 WYFKLFDQDGNGKIDKDELETIFKAIQDITRTY--DIPPE---------EIVTLIYDKIDVNGEGELTLEEFISGAKEHPDIMEMLTKMMDLSHVLEIIIKGQKKNTVK
zGCAP4 92 WYFKLFDQDGNGKIDKDELETIFTAIQDITRNR--DIVPE---------EIVALIFEKIDVNGEGELTLEEFIEGAKEHPEIMDMLKILMDLTPVLLIIVEGRQK
fuguGCAP4 92 WYFKLFDQDGNGKIDREELETIFSAIQDITRNR--DIDPE---------EIVSLIFERIDVNGEGELTLEEFIEGAKDHPDIMDTLKKIMDLTPVLVIIVEGRTG
hGCAP3 95 WYFKLYDADGNGSIDKNELLDMFMAVQALNGQQ--TLSPE---------EFINLVFHKIDINNDGELTLEEFINGMAKDQDLLEIVYKSFDFSNVLRVICNGKQPDMETDSSKSPDKAGLGKVKMK
fGCIP 94 WSFKLYDKDGDGAITRSEMLEIMRAVYKMSVVASLTKVNPMT-----AEECTNRIFVRLDKDQNAIISLQEFVDGSLGDEWVRQMLE--CDLSTVEIQKMTKHSHLPARSSRERLFHANT
fuguGCIP 95 WSFKLYDKDRDGGITRQEMLEIMQAVYKMSLAAALTRPNPLT-----AEECTNRVFARLDRDNNAIISLEEFIEGALDDDWIREMLE--CDPTTVKVERPLRRDAVLGIHG
------------ ------------
EF3 EF4 |