![]() Figure 1 of
Jiang, Mol Vis 2005;
11:143-151.
Figure 1 of
Jiang, Mol Vis 2005;
11:143-151.
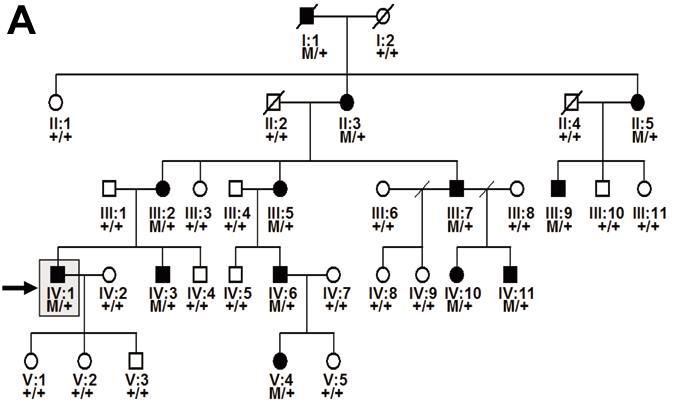
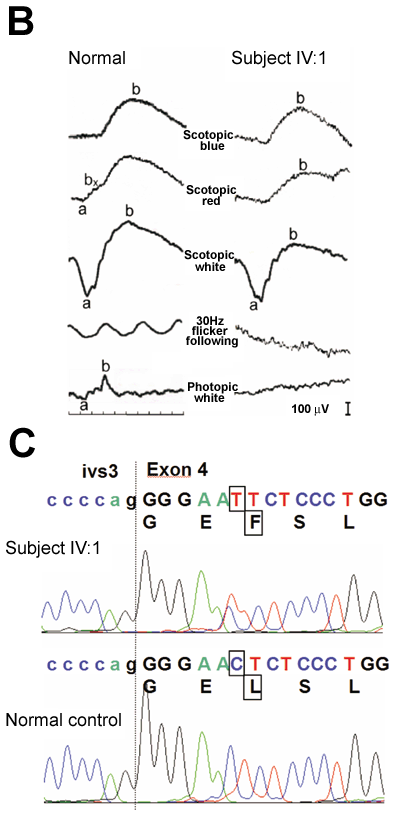
Figure 1. Identification of the GUCA1A mutation in an American Pedigree
A: Pedigree of the study family. Individuals are identified by pedigree number. Squares indicate males, circles indicate females, slashed symbols indicate deceased, solid symbols indicate affected individuals, open symbols indicate unaffected individuals, +/+ indicates two copies of wild type GUCA1A, M/+ indicates one copy of wild type GUCA1A, one copy of mutant GUCA1A. The proband analyzed in this study is identified by an arrow. B: Electroretinogram (ERG) obtained from individual IV:1. Traces obtained from the study subject are shown in the right column. Traces from a normal subject are shown in the left column for comparison. The scotopic ERGs obtained with blue, red, and white flashes are nearly normal (top three rows). The photopic ERG is non-recordable consistent with lack of cone function. 30 Hz flicker following is attenuated, but still recordable. These results are consistent with a diagnosis of cone dystrophy and are similar to electrophysiologic findings described in subjects with Y99C and E155G mutations in GUCA1A. C: Comparison of DNA sequences of the affected individual IV:1 (top) to a normal control (bottom). The proband DNA revealed a C-to-T transition (boxed) at the beginning of in exon 4 of GUCA1A, resulting in a leucine to phenylalanine change (L151F) in GCAP1 (boxed). This mutation segregated with the disease phenotype and was not found in 200 normal controls.