![]() Figure 6 of
Hemati, Mol Vis 2005;
11:1151-1165.
Figure 6 of
Hemati, Mol Vis 2005;
11:1151-1165.
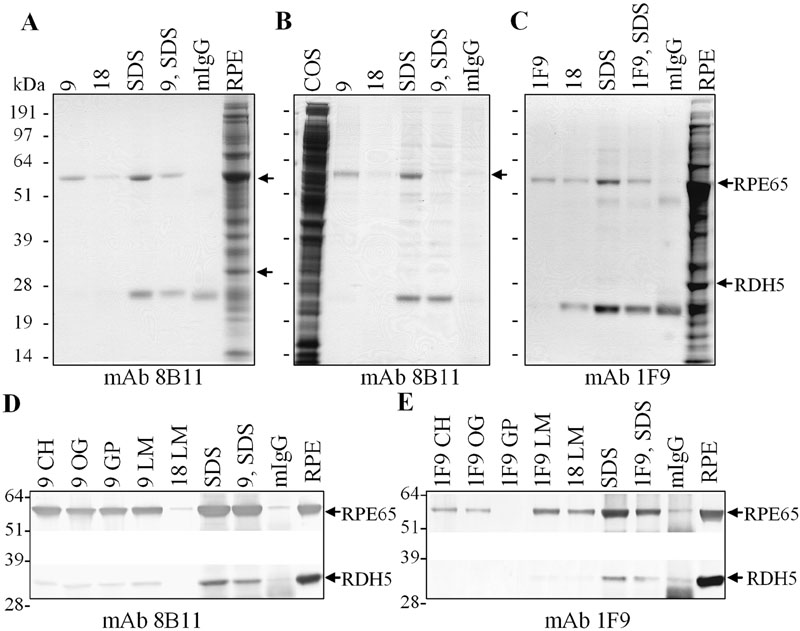
Figure 6. RPE65 immunoabsorption and peptide elution
A,B,C: Bovine RPE membranes in CHAPS (A), COS-7 cells transfected with an RPE65 cDNA expression construct in CHAPS (B), or bovine RPE membranes in laurylmaltoside (C) were incubated with mAb 8B11-Sepharose (A,B), or with mAb 1F9-Sepharose (C). Coomassie blue stained gels of proteins eluted with peptide 9 corresponding to the mAb 8B11 epitope (A,B), with the FHHINTYEDNGFLIV peptide used to generate mAb 1F9 (C), with peptide 18 corresponding to an RPE65 sequence that did not compete in ELISA's, or with SDS-sample buffer. D,E: Western analysis of bovine RPE membranes in various detergents immunoabsorbed on mAb 8B11-Sepharose (D), or on mAb 1F9-Sepharose (E), and eluted with peptides or SDS-sample buffer, as shown. Proteins were transferred to nitrocellulose and probed with mAb 8B11 (D), mAb 1F9 (E), or an antibody against RDH5 (D,E), and reactivity visualized using alkaline phosphatase coupled anti-mouse IgG. Arrows indicate the positions of RPE65 and RDH5. CH represents CHAPS; OC represents octylglucoside; GP represents Genapol; LM represents laurylmaltoside; mIgG represents nonspecific mIgG-Sepharose matrix; 9 represents KVNPETLETI; 1F9 represents FHHINTYEDNGFLIV; 9, SDS represents peptide 9 eluted matrix subsequently eluted by SDS; 1F9, SDS represents peptide 1F9 eluted matrix subsequently eluted with SDS.