![]() Figure 6 of
Gupta, Mol Vis 2005;
11:1018-1040.
Figure 6 of
Gupta, Mol Vis 2005;
11:1018-1040.
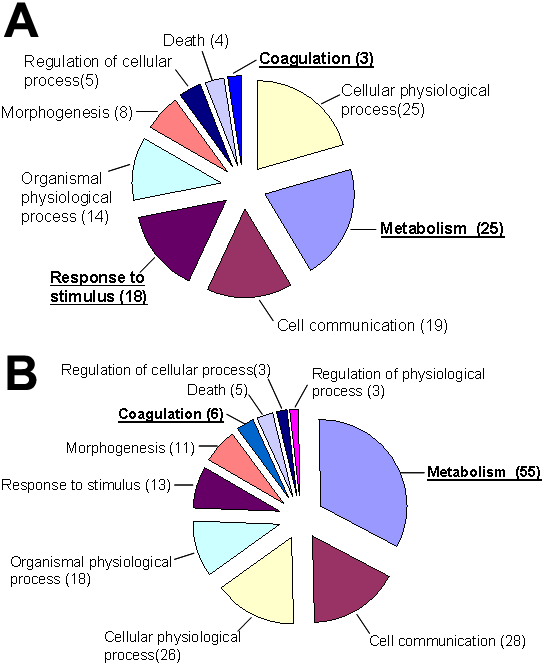
Figure 6. Cluster analysis reveals biological functions
EASE was used to statistically cluster genes that were modulated with dexamethasone (Dex) treatment compared to vehicle. The total number of genes from the filtered lists in each functional category is indicated next to the pie slice which represents the relative number of genes with modulated expression in the biological category. Some genes may be represented in more than one functional category. Not all of the genes are classified and thus do not fall into one of the functional categories represented here. Those categories with an EASE score of <0.05 are underlined in bold. A: Of the 136 genes from the 4 h Dex treatment, 77 were classified. B: Of the 86 genes from the 16 h Dex treatment, 55 were classified. Table 5 indicates the number of transcripts that were upregulated or downregulated in each of the functional categories. Although these results did not reveal a specific functional category affected by GC treatment, this is consistent with and may account for the inability of researchers, thus far, to identify a change in a biological function with GC treatment of LECs in vivo.