![]() Figure 8 of
Tasheva, Mol Vis 2004;
10:758-772.
Figure 8 of
Tasheva, Mol Vis 2004;
10:758-772.
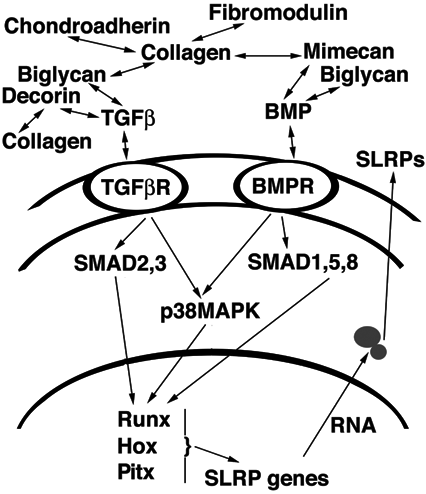
Figure 8. General model for transcriptional co-regulation of the SLRPs
General model proposing how transcription of the SLRPs may be regulated by members of the TGF-β superfamily of growth factors and by the HOX and Runx families of transcription factors. Different SLRP proteins bind TGF-β and/or bone morphogenetic protein (BMP), sequester them in the ECM without inactivation, and serve as a depot available during growth or tissue remodeling. Upon release, TGF-β and BMP interact with their respective receptors, serine-threonine kinases that signal through the SMAD family of transcriptional regulators. Other signaling pathways, such as p38 mitogen activated protein kinase (MAPK), also are activated by TGF-β and BMP signaling [78]. SMADs modulate transcription of Runx and HOX TF, and these in turn modulate transcription of SLRP genes. Some of the SLRP promoters also contain predictions of SMAD binding sites (MatInspector output). Increased/decreased expression of SLRPs that are exported back into ECM provide the feedback mechanism seen in many biological systems.