![]() Figure 3 of
Bennett, Mol Vis 2004;
10:376-382.
Figure 3 of
Bennett, Mol Vis 2004;
10:376-382.
Figure 3. Mutation analysis of GJA3
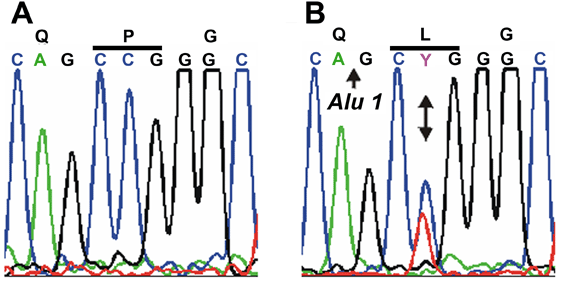
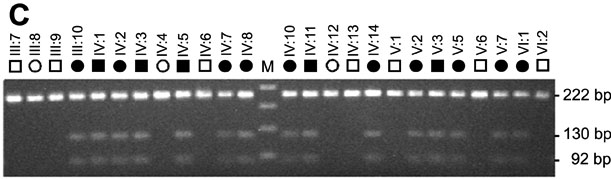
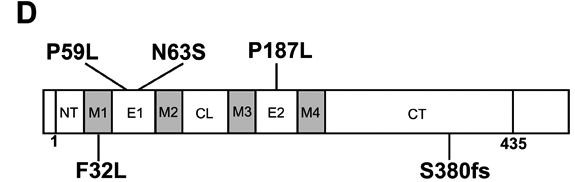
Mutation analysis of GJA3. Sequence chromatograms of wild type allele (A) showing translation of proline (CCG) at codon 59 in exon 2 and mutant allele (B) showing a C->T transition at the second base of codon 59 that substituted leucine (CTG) for proline (P59L). C: Restriction fragment length analysis on agarose gels showing gain of an Alu 1 site (5'AG/CT) that co-segregated with affected individuals heterozygous for the mutant T-allele (130 bp and 92 bp) but not with unaffected individuals homozygous for the wild-type C-allele (222 bp). The letter "M" designates the 50 bp size ladder. D: Exon organization and mutation profile of GJA3. The entire coding region (435 amino acids) of connexin46 (Cx46) is located in a single exon. Based on hydrophobicity analysis [66], Cx46 has nine structural domains including a cytoplasmic amino-terminus (NT), 4 transmembrane domains (M1-M4), 2 extracellular loops (E1-E2), a cytoplasmic loop (CL), and a cytoplasmic carboxy-terminus (CT). The relative locations, with respect to the translation start codon, of the P59L mutation and three other mutations associated with dominant cataracts in humans are indicated. E: Amino acid sequence alignment of the E1 domain (codons 42-71) from human Cx46 and homologs from other species. Dots denote identical amino acids. Cysteine residues involved in connexon hemi-channel docking are in blue. The P59L and N63S substitutions are shown in red.
E:
L S
| |
59 63
| |
Human Cx46 EDVWGDEQSDFTCNTQQPGCENVCYDRAFP
Rat Cx46 .E............................
Mouse Cx46 .E............................
Bovine Cx44 .E............................
Sheep Cx44 .E............................
Chicken Cx56 .E........................K...
Zebrafish Cx48.5 .E........................E...
|