![]() Figure 3 of
Zhang, Mol Vis 2004;
10:884-889.
Figure 3 of
Zhang, Mol Vis 2004;
10:884-889.
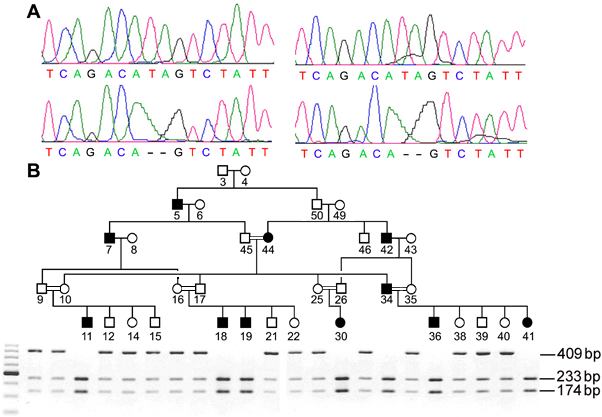
Figure 3. CNGA1 sequence changes in affected individuals
A: Top shows forward (left) and reverse (right) sequencing results of normal exon 8. Bottom shows forward (left) and reverse (right) sequences of exon 8 for patients number 11 showing homozygous TA deletion. B: Pedigree of family 61039 with agarose gel of HpyCH4III digestion products shown beneath each individual. HpyCH4III digestion of exon 8 PCR products (409 bp/wt and 407 bp/mutant). The TA deletion creates a HpyCH4III site. The 409 bp wild type fragments were not cut by HpyCH4III but the 407 bp mutant fragments were cut into two fragments: 233 bp and 174 bp. Heterozygous carriers had 409 bp, 233 bp, and 174 bp bands. Gel patterns for family members are shown directly below that individual in the pedigree. DNA from individuals 3-8, 26, 42-45, 49, and 50 were not loaded on the gel. Heterozygous carriers have 409 bp, 233 bp, and 174 bp bands. All unaffected individuals examined in the family are heterozygous carriers who have all three bands. Patients homozygous for the mutation have only the two lower bands (233 bp, 174 bp) without the upper band at 409 bp. Unaffected individuals not carrying the mutation show a single upper band of 409 bp (not shown in this figure).