A Polymorphic Trinucleotide Repeat at DXS8170 in the Critical Region of X-Linked Retinitis Pigmentosa Locus RP3 at Xp21.1
Ricardo Fujita1*, Mark Blumberg1, David Anderson1, Patricia Forsythe1, Christina McHenry1, Denise Yan1, Teresa L. Yang-Feng2, Paul A. Sieving1, and Anand Swaroop1,3*
1Departments of Ophthalmology and 3Human Genetics, W.K. Kellogg Eye Center, University of Michigan, Ann Arbor, MI 48105, USA.
2Department of Genetics, Yale University School of Medicine, New Haven, CT 06510.
*Corresponding authors email: swaroop@umich.edu and rfujita@umich.edu
Retinitis Pigmentosa (RP) constitutes a group of genetically heterogeneous diseases, with at least 15 different mapped genetic loci associated with varying phenotypes. Probably the most complex scenario is presented by the X-linked forms of RP (XLRP), for which four loci are clustered on the short arm of the X chromosome (RP2, RP3, RP6, and RP15) between Xp11.23 and Xp22.13 (4-6). In addition, genetic loci for X-linked cone-rod dystrophy (COD1), congenital stationary night blindness (CSNB), and Åland Island eye disease (AIED) have been localized in this chromosomal region (5, 6). It is possible that these diseases are caused by allelic variants of the XLRP genes. The XLRP locus RP3 has been genetically localized between OTC and DXS1110 , which are currently the closest proximal and distal flanking markers respectively, and are separated by approximately 500 kb of DNA. The proximal chromosomal breakpoint in the patient BB (who had a cytogenetically visible deletion and suffered from XLRP in addition to DMD, CGD and the McLeod Syndrome) physically pinpoints the RP3 gene to a region of 40-50 kb proximal to the marker DXS1110 (5). In spite of an extensive search, the RP3 gene has not yet been identified in this region. In our continuing efforts to clone the RP3 gene, we have isolated about 450 kb of genomic DNA spanning DXS1110 and the BB proximal breakpoint in a yeast artificial chromosome (YAC) clone and cosmid subclones (2). We have searched for new polymorphic markers in the cosmids located between DXS1110 and OTC . Here we report a new polymorphic trinucleotide repeat which will assist in refining the genetic map position of RP3 and other X-linked retinopathies in this region and will be valuable for prenatal diagnosis and carrier detection.
Results
The 55B YAC clone of ~450 kb (DXS1172 ), was isolated by screening the Washington University YAC library (1) with primer sequences derived from the marker p55.5 (DXS140 ) (2). Physical mapping of the 55B YAC clone showed that it spans the proximal breakpoint of the patient BB and includes the XK , CYBB , and TCTEX-1L genes (3, 5). A cosmid sub-library, constructed from this YAC, was screened with a 32P-labeled probe generated from a pool of ten trinucleotide repeat oligomers. A 0.66 kb PstI fragment (MB1) was isolated from one of the positive cosmid clones (c3.19), which is located ~160 kb proximal to the DXS1110 locus. The complete sequence of the MB1 subclone (locus name DXS8170 ; GenBank accession # U32657) revealed an imperfect trinucleotide repeat: (GAG)2 GAA (GAG)5 AAG (GAG)3 A (TGG)2 TGC (TGG)2 TGATNGGGGGAAG (GAG)5 (AAG)2 (GAG)2. Polymorphism
The primers flanking the repeat amplified a segment of 179 nucleotides in the MB1 clone. The amplified product demonstrated a two-allele trinucleotide polymorphism (179 and 182 bp) in the human genomic DNA.
PCR Conditions
Sequences MB1-B4 (5' TGG CTC CGA AAC TCA GGA ATC 3') and MB1-F1 (5' TCA GAT GGA TCT TGG AGT TGG 3'), which flanked the microsatellite, were used to synthesize the PCR primers. Amplification was performed in a 20 microliter reaction containing 50 ng of DNA, 10 ng of each primer (one of these labeled with 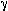 -32P-ATP using T4 polynucleotide kinase), 100 micromolar each dNTP, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1 mM MgCl2 and 0.5 units of AmpliTaq (Perkin-Elmer). Reaction was carried out for 35 cycles of 45 sec. at 94oC, 45 sec. at 57oC and 45 sec. at 72oC. Two microliters of the reaction was loaded on a 6% polyacrylamide-50% urea denaturing gel which had been pre-run for 30 min.
-32P-ATP using T4 polynucleotide kinase), 100 micromolar each dNTP, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1 mM MgCl2 and 0.5 units of AmpliTaq (Perkin-Elmer). Reaction was carried out for 35 cycles of 45 sec. at 94oC, 45 sec. at 57oC and 45 sec. at 72oC. Two microliters of the reaction was loaded on a 6% polyacrylamide-50% urea denaturing gel which had been pre-run for 30 min.
Frequency/Chromosomal Localization/Inheritance
Two alleles of 179 and 182 bp were observed in 101 independent chromosomes (29 females and 43 males). The frequency of the 179 bp allele was 26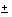 8.6% with a 95% confidence interval. The 55B YAC clone and c3.19 cosmid were localized to Xp21.1 by fluorescence in situ hybridization (2). Thirteen out of 29 females were heterozygous (45%). Most of these women were caucasian. An X-linked pattern of segregation was observed in two large three-generation and 20 nuclear XLRP families. The location of DXS8170 on the X-chromosome was confirmed by the heterozygosity observed only in females of a sample that included 29 females and 43 males. Figure 1 shows the X-linked segregation of DXS8170 observed in two XLRP families.
8.6% with a 95% confidence interval. The 55B YAC clone and c3.19 cosmid were localized to Xp21.1 by fluorescence in situ hybridization (2). Thirteen out of 29 females were heterozygous (45%). Most of these women were caucasian. An X-linked pattern of segregation was observed in two large three-generation and 20 nuclear XLRP families. The location of DXS8170 on the X-chromosome was confirmed by the heterozygosity observed only in females of a sample that included 29 females and 43 males. Figure 1 shows the X-linked segregation of DXS8170 observed in two XLRP families.
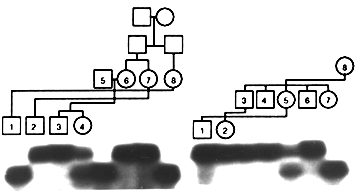
Figure 1: The segregation of DXS8170 alleles in two XLRP families showing an X-linked pattern of inheritance. A part of the pedigrees is shown above the lanes.
Discussion
DXS8170 is located in the critical region for RP3, which is the best defined genetic locus among retinal diseases mapped to the short arm of the X-chromosome. Physical mapping studies demonstrate that DXS8170 is between DXS1110 and OTC, currently the closest flanking markers for RP3. If the proximal BB breakpoint is an indicator for the gene, DXS8170 is closer to RP3 than OTC on the proximal side. Alternatively, it is also possible that RP3 lies away from the proximal BB breakpoint. In this scenario, DXS8170 will be closer to the gene than DXS1110. The genetic analysis of RP3 families with the DXS8170 marker will localize its position with respect to the disease locus. Since a cluster of several distinct retinal dystrophies reside within the 20-25 cM region at the short arm of X-chromosome (RP2, RP3, RP6, RP15, COD1, CSNB1 and AIED), this new marker will assist in refining the position of potential meiotic recombinations, thereby contributing to the identification of these disease genes.
Acknowledgements
We thank Ms. Eve Bingham for the gift of normal human DNA samples. This work was supported by grants from the National Institutes of Health (EY07961) and the Foundation Fighting Blindness, Hunt Valley, MD. We also acknowledge NIH grants EY07003 (CORE) and M01-RR00042 (General Clinical Research Center).
REFERENCES
1. Brownstein BH, Silverman GA, Little RD, Burke DT, Korsmeyer S J, Schlessinger D, and Olson MV (1989). Isolation of single copy human genes from a library of yeast artificial chromosome clones. Science 244: 1348-1351. 2. Fujita R, Ayon D, Skolnick C, Yang-Feng T, and Swaroop A (1993). Isolation, characterization and subcloning of YACs spanning OTC, CGD, RP3 and McLeod phenotype loci. Cytogenet Cell Genet 64: 177.
3. Ho M, Chelly J, Carter N, Danek A, Crocker P, and Monaco AP (1994). Isolation of the gene for McLeod syndrome that encodes a novel membrane transport protein. Cell 77: 869-880.
4. McGuire RE, Sullivan LS, Blanton SH, Church MW, Heckenlively JR, and Daiger SP (1995). X-linked dominant cone-rod degeneration: Linkage mapping of a new locus for retinitis pigmentosa (RP15) to Xp22.13-p22.11. Am J Hum Genet 57: 87-94.
5. Willard HF, Cremers F, Mandel JL, Monaco AP, Nelson DL, and Schlessinger D (1994). Report of the fifth international workshop on human X chromosome mapping 1994. Cytogenet Cell Genet 67: 295-358.
6. Sixth X Chromosome Workshop, Banff, Alberta, Canada; June 1995
 Return to Molecular Vision homepage
Return to Molecular Vision homepage
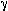 -32P-ATP using T4 polynucleotide kinase), 100 micromolar each dNTP, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1 mM MgCl2 and 0.5 units of AmpliTaq (Perkin-Elmer). Reaction was carried out for 35 cycles of 45 sec. at 94oC, 45 sec. at 57oC and 45 sec. at 72oC. Two microliters of the reaction was loaded on a 6% polyacrylamide-50% urea denaturing gel which had been pre-run for 30 min.
-32P-ATP using T4 polynucleotide kinase), 100 micromolar each dNTP, 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 1 mM MgCl2 and 0.5 units of AmpliTaq (Perkin-Elmer). Reaction was carried out for 35 cycles of 45 sec. at 94oC, 45 sec. at 57oC and 45 sec. at 72oC. Two microliters of the reaction was loaded on a 6% polyacrylamide-50% urea denaturing gel which had been pre-run for 30 min. 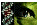
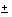 8.6% with a 95% confidence interval. The 55B YAC clone and c3.19 cosmid were localized to Xp21.1 by fluorescence in situ hybridization (2). Thirteen out of 29 females were heterozygous (45%). Most of these women were caucasian. An X-linked pattern of segregation was observed in two large three-generation and 20 nuclear XLRP families. The location of DXS8170 on the X-chromosome was confirmed by the heterozygosity observed only in females of a sample that included 29 females and 43 males. Figure 1 shows the X-linked segregation of DXS8170 observed in two XLRP families.
8.6% with a 95% confidence interval. The 55B YAC clone and c3.19 cosmid were localized to Xp21.1 by fluorescence in situ hybridization (2). Thirteen out of 29 females were heterozygous (45%). Most of these women were caucasian. An X-linked pattern of segregation was observed in two large three-generation and 20 nuclear XLRP families. The location of DXS8170 on the X-chromosome was confirmed by the heterozygosity observed only in females of a sample that included 29 females and 43 males. Figure 1 shows the X-linked segregation of DXS8170 observed in two XLRP families.